3.6.2 The JAK–STAT pathway
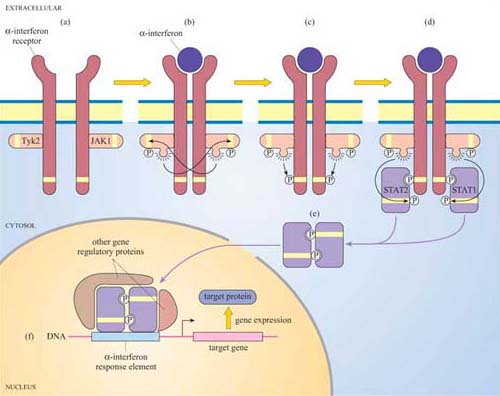
Another important protein kinase pathway is the JAK–STAT pathway. Cytokines (Section 2.2), are frequently used for signalling between cells of the immune system. Cytokine-induced signal transduction cascades are often direct pathways to the nucleus for switching on sets of genes. Janus kinases (JAKs, named after the two-faced Roman god) are a particular group of tyrosine kinases that associate with cytokine receptors. When cytokines such as α-interferon bind to their receptor, JAKs (denoted as JAK1, Tyk2, etc.) associated with different receptor units cross-phosphorylate each other on tyrosine residues (Figure 41). The receptors are then phosphorylated by JAKs, creating phosphotyrosine-docking sites for SH2-containing proteins, in this case a specific group of proteins called STATs (signal transducers and activators of transcription).
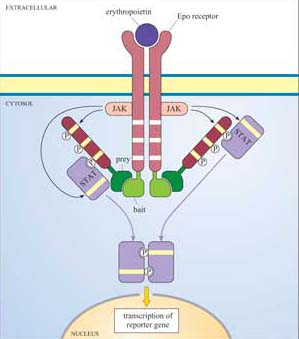
Box 4 Detecting protein–protein interactions using the MAPPIT system
The mammalian protein–protein interaction trap (MAPPIT) system uses the principle, instead of the genes of interest being fused to transcriptional activator domains, they are fused to components of the JAK–STAT signalling pathway (Figure 42). Using recombinant DNA technology, the bait protein domain is fused to a mutant erythropoietin (Epo) receptor, which has JAK binding sites but lacks STAT binding sites. The prey protein is fused to a protein fragment that has several potential STAT binding sites. If there is no interaction between bait and prey, JAKs cross-phosphorylate each other on stimulation with Epo, but STAT is not recruited as there are no available docking sites on the receptor. If there is an interaction between bait and prey, the protein fragment becomes tyrosine-phosphorylated by JAKs on receptor activation, enabling STAT to bind to it. STAT is then in the vicinity of JAK, which can access and phosphorylate the STAT. Then STAT can dimerize, translocate to the nucleus and induce transcription of a reporter gene which can be easily quantified.
In turn, the STATs are phosphorylated by the JAKs, which causes the STATs to dissociate from the receptor and instead bind to each other by means of their SH2 domains. STAT dimers translocate directly to the nucleus, where they bind to other gene regulatory proteins and to response elements in target genes.
Can you recall any other signalling pathway that has also has a direct route to the nucleus?
The nuclear hormone receptor signalling pathway, where the intracellular receptor binds the ligand (for example, a steroid) and translocates to the nucleus, where it binds to hormone response elements (Figure 27).
The JAK–STAT pathway has recently become the basis for the development of techniques for the study of protein–protein interactions (Section 1.5).
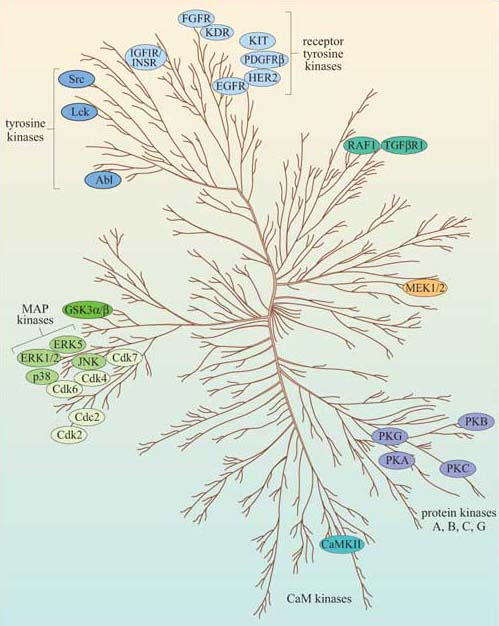
Now that we have described the major protein kinases, we can look at them all together, and see what features they have in common. Figure 43 is a dendrogram showing how the various subgroups of protein kinases in humans are related to each other both functionally and structurally. Despite their different subcellular localizations, activation mechanisms and substrates, protein kinases have remarkable similarity of structure within their kinase domain.