2.2 Inhibitors of protein synthesis
You learned in Activity 1 that cells synthesise new proteins in ribosomes which are made up of one large and one small subunit. These subunits differ structurally and chemically between prokaryotic and eukaryotic ribosomes (Figure 5). This provides antibiotic targets in the bacterial pathogen which are not present in the host cells.
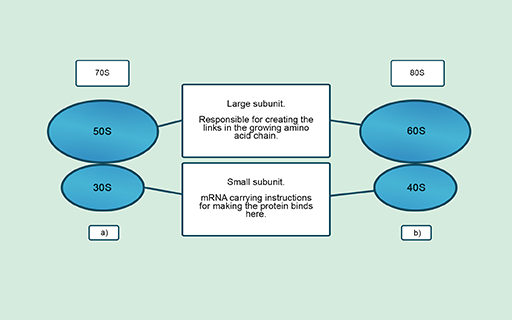
Figure 5 Ribosome structure in (a) prokaryotes and (b) eukaryotes. The Svedberg unit (S) indicates the size, shape and density of each subunit.
| Ribosomal target | Outcome | Antibiotic class | Structure | Example drug |
|---|---|---|---|---|
| Small (30S) subunit | Errors give rise to faulty proteins that disrupt the cell membrane | Aminoglycosides | All contain amino sugar substructures (red)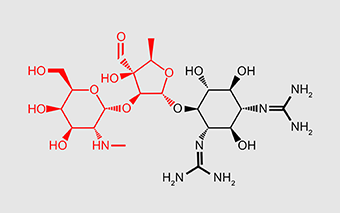 | Streptomycin |
| Large (50S) subunit | First steps of protein synthesis (initiation) are impaired and bacteria cannot grow and divide | Oxazolidines | All contain 2-oxazolidone (red) somewhere in their structure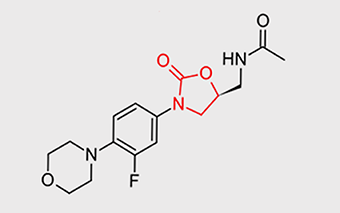 | Linezolid |
(OpenStax College Microbiology, n.d.)
