4.4 Ribosomes: the sites of protein synthesis
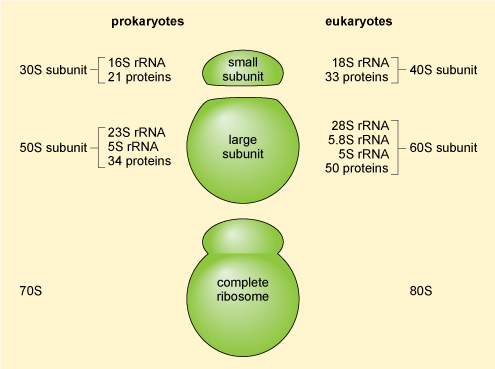
The mRNA molecules exported from the nucleus are translated into protein in the cytoplasm by the ribosomes (which are RNA-protein complexes, not organelles). Evidence suggests that some mRNA molecules leaving the nucleus are directed to specific sites in the cell before they are translated. Ribosomal structure is similar in prokaryotic and eukaryotic cells. Ribosomes are organised into a large subunit and a small subunit, as shown in Figure 15 (prokaryotic ribosomes contain over 50 different types of protein and those of eukaryotic cells contain more than 80 different types of protein). The large subunit contains two different ribosomal RNAs in prokaryotes and three in eukaryotes, while the small subunits contain one ribosomal RNA'.
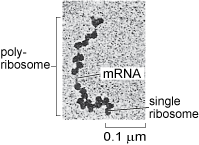
Ribosomes occur both 'freely' in the cytosol, i.e. not attached to any organelle (Figure 16), or attached to the membrane of the rough endoplasmic reticulum in eukaryotic cells (Figure 1a and b). Cytosolic proteins and proteins that are destined for the nucleus, mitochondria, chloroplasts and peroxisomes (you will learn about these other organelles later in this course) are synthesised by the free ribosomes in the cytosol. Cytosolic proteins, such as cytoskeleton components, are simply released into the cytosol once their synthesis is complete, but many other proteins are delivered to specific locations by a complex system of protein targeting or protein sorting. A signal sequence, a characteristic short sequence of particular amino acids in the protein itself, acts rather like a postal address that targets the protein for import into the correct organelle. Nuclear proteins, for example, contain a nuclear localisation sequence and are recognised by proteins that transport them to the nuclear membrane and through the nuclear pore. Proteins destined for mitochondria, chloroplasts or peroxisomes also have a characteristic signal sequence directing them to the appropriate organelle. This 'tagging' of polypeptides by signal sequences comprising specific amino acids is an important general mechanism used by all cells to recognise and deliver proteins to specific sites in the cell. Prokaryotic cells lack organelles, but they use a similar mechanism to target proteins to the cell membranes, cell wall, or to various types of inclusions in the cytosol.
As you will learn in the next section, lysosomal proteins (Section 4.9), and proteins that are destined for export from the cell, or to reside in the cell membrane, are translated by ribosomes that are attached to the endoplasmic reticulum.