A great deal of experimental data generated by scientists is now stored online in publicly available databases. Because of this, it can now be used (‘interrogated’) in large biological studies to uncover clues about the functions of genes, other genetic components, proteins, and cell pathways that are important in cancers and other diseases. This process of finding out data is called data mining. Bioinformaticians are scientists who specialise in developing and using specialist software tools to do just this; to analyse and interpret combinations of big data sets generated in biological studies.
The creation of new analysis tools for working with this type of data is therefore becoming increasingly important. It allows scientists to fully make use of the information available, and allows them to compare and analyse data generated from different sources. It can be used to find new links between genes and disease that may be responsible for diseases such as cancer. Armed with this information we can work towards the development of personalised screening tools such as blood tests, or specific treatment regimens that would increase the chances of an individual responding to a given cancer therapy. Bioinformatics is a therefore a cornerstone of personalised medicine – allowing large data sets to be used to customise medical treatment
Delving into DNA
DNA consists of 4 basic building blocks called nucleic acid bases: adenosine (A) thymine (T) cytosine (C) and guanine (G) and are arranged in a particular order (sequence) so that when they are read by the cell’s machinery (RNA polymerase) they ‘code’ (have instructions) for particular products, proteins, to be produced. For example the protein pigments that give eye colour. However these sequences or some of these nucleic acid bases, are slightly different between individuals called variants, which can be inherited or changed during a person’s lifetime. These variants can span small regions of DNA and can be from just one DNA base to much larger regions, hundreds of bases. Certain sequences are more common in certain populations, which sometimes gives us information about increased risks of particular diseases within that population. For example in Caucasian (White) populations there is a higher risk of breast cancers.
The increase in the amount and accessibility of ‘big data’ allows for the analysis of genetic material from entire populations. in databases for example: the National Centre for Biotechnology Information (NCBI) dbSNP and dbVar databases, the Encyclopaedia of Coding Elements (ENCODE & UCSC), and the Catalogue of Somatic Mutations in Cancer (COSMIC) are just a few. Information stored in these databases are the deoxyribonucleic acid (DNA) sequences along with information about the traits or characteristics of thousands of human samples.
Bioinformatics for cancer
Using this genetic data and combining it with data from other studies provides greater power to find their functions and links that may underlie cancers. For example, there are a number of identified and confirmed breast cancer causing variants that are associated with certain genes, (segments of a DNA sequence that causes the production of a certain protein). These genes are involved in a number of actions such as cell pathways and gene regulatory processes that lead to the normal controlled growth and eventual death of a cell. However when variants occur these normal actions can be disrupted and lead to uncontrolled cell growth (cancerous) e.g. BRCA1 (breast cancer 1) and BRCA2 (breast cancer 2) DNA repair genes in inherited breast cancer.
Bioinformaticians can interrogate the large data sets using publically available online tools to identify these variants and determine whether there are genes or non-coding regions of DNA underlying multiple pathways or processes in cancers. Some of these tools are built into databases directly, to help scientist carry out such analyses. Tools such as R and R’s Bioconductor are programs that allow users to analyse data of DNA sequences, traits, and other genetic components from the databases (using basic computer coding) to determine the role variants play in breast cancer.
By identifying the variants in certain populations, bioinformaticians hope to identify or predict individuals that may have an increased risk of developing breast cancer during their lifetime and whether other cancers are related to the underlying mechanisms.
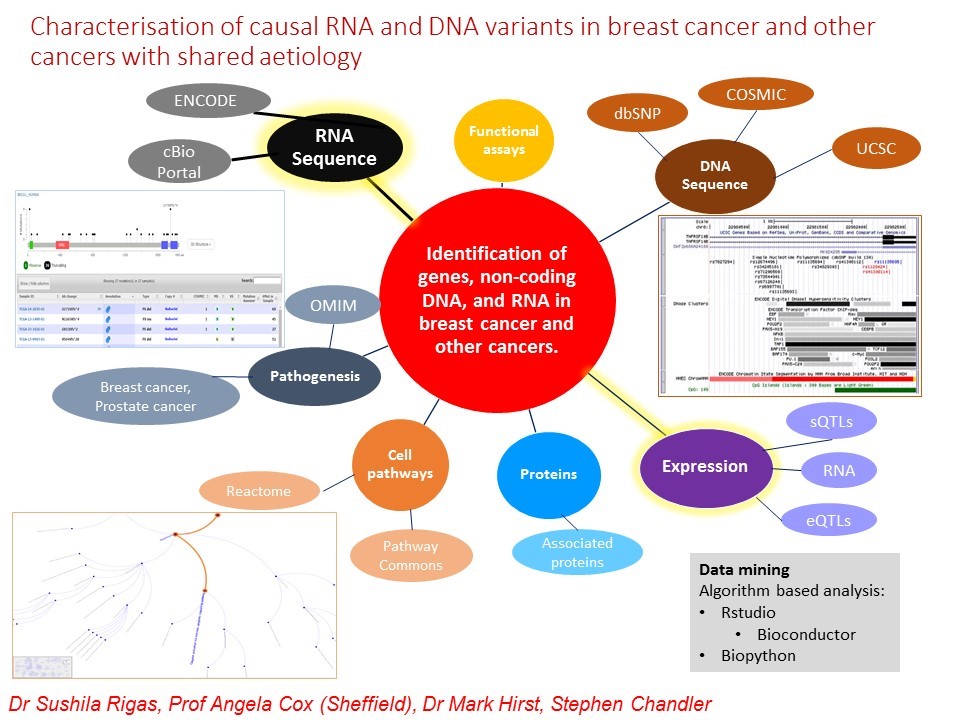 Figure 1: Approaches and databases used to identify DNA variants, genes, and other genetic components, pathways, and processes. The inner circles attached to the red circle are genetic components, processes, and pathways that will be investigated. Each outer circle gives the databases or characteristics that can be investigated with images of the use of some databases
Figure 1: Approaches and databases used to identify DNA variants, genes, and other genetic components, pathways, and processes. The inner circles attached to the red circle are genetic components, processes, and pathways that will be investigated. Each outer circle gives the databases or characteristics that can be investigated with images of the use of some databases



Rate and Review
Rate this article
Review this article
Log into OpenLearn to leave reviews and join in the conversation.
Article reviews