5.2 Determining the subtype of influenza
Immunofluorescence
Confirmation of a case of influenza is usually achieved by performing tests on some of the inoculated cultured cells using reference fluorescent-labelled antisera provided by the WHO. A reference antiserum is a sample serum known to contain antibodies specific for the molecule to be assayed (in this case, haemagglutinin or neuraminidase). These antisera are prepared using purified haemagglutinin and neuraminidase and are monospecific, each antibody reacting only with one epitope e.g. H1 or H3.
For the test, an antibody is added to a sample of inoculated cells. Following a wash step, if the antibody remains bound to the cells then they fluoresce under appropriate illumination, indicating the presence of viral antigen on the cell surface. This diagnostic technique can identify influenza virus on infected cells in as little as 15 minutes. A positive result not only confirms the RT-PCR data, but gives additional information on the subtype of the virus.
Further PCR analyses
Standardised RT-PCR protocols exist to look for the presence of different haemagglutinin and neuraminidase subtypes, chiefly H1, H3, H5, N1 and N2. (Poddar, 2002). If the PCR analysis indicates a dangerous strain of influenza A e.g. H5N1, then it is instantly sent to a WHO reference laboratory for further tests.
Haemagglutination assays
Influenza has haemagglutinins protruding from its viral envelope, which it uses to attach to host cells prior to entry. These substances form the basis of a haemagglutination assay, in which viral haemagglutinins bind and cross-link (agglutinate) red blood cells added to a test well, causing them to sink to the bottom of the solution as a mat of cells. If agglutination does not occur, then the red blood cells are instead free to roll down the curved sides of the tube to form a tight pellet.
A related test called a haemagglutination-inhibition assay (HAI), incorporates antibodies against different subtypes of viral haemagglutinin. The antibodies bind and mask the viral haemagglutinin, preventing it from attaching to and cross-linking red blood cells.
A HAI assay can be set up in one of two ways: either a known reference antibody is added to an unknown virus sample, or known reference viral haemagglutinin is added to a sample of patient serum containing antibodies against influenza. This second version of the HAI assay can therefore be used long after the infection has passed, when virions are no longer present.
Haemagglutination and HAI assays have the advantage that they are simple to perform and require relatively cheap equipment and reagents. However, they can be prone to false positive or false negative results, if the sample contains non-specific inhibitors of haemagglutination (preventing agglutination) or naturally occurring agglutinins of red blood cells (causing agglutination).
If a confirmed influenza A isolate reacts weakly or not at all in HAI then this indicates an unknown variant of influenza A and the sample is immediately sent to a WHO reference laboratory for further tests.
Neuraminidase inhibition assay
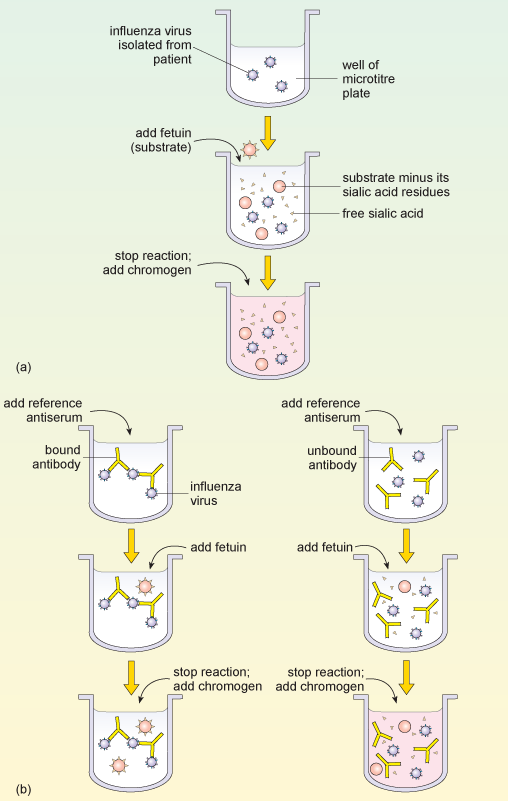
Typing influenza isolates in terms of their neuraminidase makes use of the enzyme activity of this glycoprotein. The neuraminidase inhibition assay is performed in two parts. The first part determines the amount of neuraminidase activity in a patient influenza sample, as outlined in Figure 9a. A substrate (called fetuin) that is rich in sialic acid residues is added to a sample of the influenza virus, and the viral neuraminidase enzyme cleaves the substrate to produce free sialic acid.
Addition of a substance that inactivates the neuraminidase stops the reaction, and a chromogen (a colourless compound that reacts to produce a coloured end-product) that turns pink in the presence of free sialic acid is added. The intensity of the pink colour is proportional to the amount of free sialic acid and can be measured using a spectrophotometer.
This assay of neuraminidase activity allows the appropriate amount of virus sample to be determined, and this quantity is then used in the second part of the assay. If too much or too little virus is used, the resulting changes, and therefore the neuraminidase, may be undetectable.
In the second part of the assay (Figure 9b), viral samples from the patient are incubated with anti-neuraminidase reference antisera. Each of the reference antisera used for this test has antibodies that bind one particular neuraminidase variant, e.g. N1 or N2.
How can these antisera be used to type the neuraminidase variant?
If the antibodies bind the neuraminidase in the patient’s sample they inhibit its activity. This means that the patient’s neuraminidase cannot cleave the sialic acid from its test substrate fetuin, so no colour change will occur when the chromogen is added. Conversely, if the antibodies in the reference antiserum do not bind the neuraminidase in the patient’s sample, then the enzyme will remain uninhibited and the pink colour will be produced as before.