5.2 Biological targets of cisplatin
As it is clear that the DNA in the cell is being targeted by the cisplatin, it is useful to consider which parts of the DNA might be preferentially bound by the metal ion.
You can refresh your memory about the molecular structure and function of DNA in Box 1.
Box 1 DNA
DNA, or deoxyribonucleic acid, is a biopolymer composed of repeat monomers known as nucleotides. Each nucleotide consists of:
- a phosphate group
- a sugar molecule
- a nitrogen-containing base.
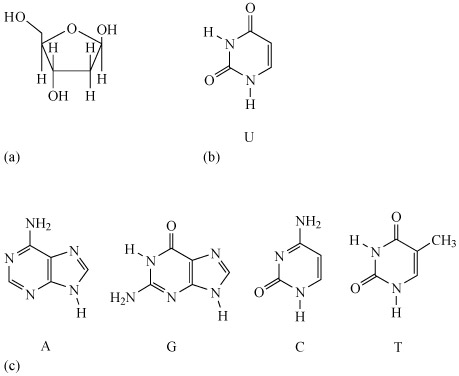
The sugar is deoxyribose (Figure 17a).
There are four different bases in DNA: adenine, guanine, cytosine and thymine, usually abbreviated to A, G, C and T, respectively. Their structures are shown in Figure 17b. A fifth base, called uracil, U (Figure 17c), usually takes the place of thymine in RNA and differs from thymine by lacking the methyl group on its ring.
DNA has a double-helix structure, which gives it stability.
The strand of alternating phosphate groups and sugars with ester linkages forms the sugar–phosphate backbone of each strand, and the bases protrude out from this towards the other strand of the helix. Along the length of the polynucleotide chain, each base makes a specific pairing (Watson–Crick pairing) with a corresponding base in the other polynucleotide chain with hydrogen-bonding: T (thymine) pairs only with A (adenine), and C (cytosine) pairs only with G (guanine).
The pairs of complementary bases are thus T and A, and C and G, as Figure 18 shows for a portion of a DNA molecule. These are by far the largest known molecules in living organisms, some containing millions of nucleotides.
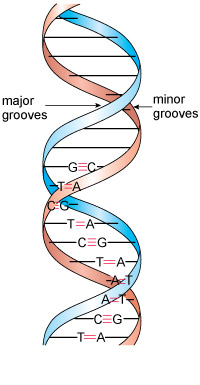
Inspection of the double-helix structure reveals two grooves (see Figure 18): the major groove and the minor groove, with approximate widths of 220 and 120 pm, respectively.
It is important to note that the bases are more exposed in the major groove and are therefore more accessible to other molecules, and this is where most reactions take place.
DNA can take a number of different conformations, of which three are found in nature, labelled as A-, B- and Z-DNA. Of these, B-DNA is the form most commonly found in cells.
The likely targets for the platinum ion would appear to be the nucleotide bases, and you will consider the chelating abilities of each base in turn.
But first, return to the work of Professor Lippard by watching the following video.
Transcript: Video 8 The cisplatin story: Part 3. (1:38 min)
NARRATOR: DNA is the complex molecule inside all living cells, shown here in a very simplified form. Like all cells, cancer cells grow by reproducing the DNA. And the assumption had to be that, somehow, cisplatin’s anticancer properties had to do with its interference in this process.
STEPHEN LIPPARD: Every time the cell divides it has to reproduce its DNA, completely and accurately. And, in order for it to function, it has to take the message that is encoded in the DNA, transcribe it into RNA and ultimately translate that into proteins. We know that platinum blocks both of these processes – replication and transcription.
NARRATOR: The vital portions of DNA are the centre groups. These are known as bases and they hold together the two strands of the double helix. The bases are adenine, thymine, cytosine and guanine. This is the chemical formula for guanine. What is important here is the nitrogen labelled as N7. It’s important because NMR was able to show that cisplatin could bond to the nitrogen at this site on the guanine base. This early work showed that most of the cisplatin formed what is called an adduct by linking to two neighbouring guanine bases on the same strand of the DNA through the N7 atom. We call this an intrastrand adduct because the bonding is all on the same strand.
-
From the NMR studies, to which base does the cisplatin appear to bind preferentially?
-
The cisplatin binds to the guanine base, coordinated with the nitrogen labelled as N7.
Theoretical and experimental studies have shown that the N7 atom on guanine (imidazole) is the most electron-rich centre. (Remember that atom numbering starts at the functional group, as shown in Structure 4.)
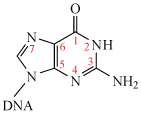
The monohydrated cisplatin reacts with DNA to form adducts, mostly forming Pt–N bonds to guanine N7, as shown in Structure 5.
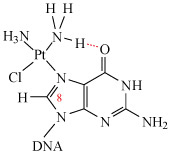
The Pt–G(N7) bond is very stable and can only be broken by a strong nucleophile (e.g. ). Hydrogen-bonding may also stabilise the adduct (as shown in Structure 4).
This adduct can be readily detected using spectroscopy: the resonance of the C8–H (shown in Structure 5) in guanine is a singlet at δ = 7.8 ppm. However, when guanine is complexed to cisplatin at N7, this singlet shifts to δ = 8.8 ppm and satellites are also observed.
This large shift means that becomes a very useful tool in elucidating the structures of these more complex systems – along with the crucial technique of X-ray crystallography, as you will see in the next section.