Use 'Print preview' to check the number of pages and printer settings.
Print functionality varies between browsers.
Printable page generated Friday, 21 November 2025, 6:37 AM
Isolating and identifying bacteria (human health)
Introduction
In this course you will learn about microbiology techniques for identifying bacteria, focusing on
Basic knowledge of concepts such as bacterial growth and bacterial structure is assumed. If you are unfamiliar with these concepts or would like a refresher course, you might want to look back at the Introducing antimicrobial resistance course or this Microbiology textbook (OpenStax, 2021). Information about the GLASS programme can be found in the Introducing AMR surveillance systems course.
After completing this course, you will be able to:
- know where samples are obtained from and reflect on the process by which bacterial samples are processed in your workplace
- describe the principles of laboratory tests used to isolate and identify key bacterial pathogens in human health, which are the focus of the GLASS programme
- know when and why advanced testing such as mass spectrometry and automated systems are used
- know the importance of procedures designed to ensure the quality of laboratory work relating to isolating and identifying bacteria in your workplace.
In order to achieve your digital badge and Statement of Participation for this course, you must:
- click on every page of the course
- pass the end-of-course quiz
- complete the course satisfaction survey.
The quiz allows up to three attempts at each question. A passing grade is 50% or more.
When you have successfully achieved the completion criteria listed above you will receive an email notification that your badge and Statement of Participation have been awarded. (Please note that it can take up to 24 hours for these to be issued.)
Activity 1: Assessing your skills and knowledge
Before you begin this course, you should take a moment to think about the learning outcomes and how confident you feel about your knowledge and skills in these areas. Do not worry if you do not feel very confident in some skills – they may be areas that you are hoping to develop by studying these courses.
Now use the interactive tool to rate your confidence in these areas using the following scale:
- 5 Very confident
- 4 Confident
- 3 Neither confident nor not confident
- 2 Not very confident
- 1 Not at all confident
This is for you to reflect on your own knowledge and skills you already have.
1 Principles of sampling and specimen collection
In this section we introduce the basics of sampling and how to collect and process clinical specimens.
1.1 Origins of samples
Clinical samples come from a range of different settings, such as:
- secondary and tertiary care patients in hospital
- community patients seen in clinics
- specialist community clinics and outpatient departments, for example tuberculosis (
TB ) or sexual health services.
Activity 2: Types of sample
Make notes on the types of samples that are sent to your microbiology laboratory. Compare your response with the sample answer.
Discussion
Microbiology samples include the following:
- Swabs, for example from wounds, throats, eyes and ears
- Genital (cervical and urethral) samples
- Urine
- Blood
- Other normally sterile fluids, such as cerebrospinal fluid (
CSF ), synovial fluid - Stools
- Sputum – for TB and sometimes other respiratory pathogens
- Pus and tissue from sterile and non-sterile sites.
Clinical samples are generally sent to microbiology laboratories to guide antimicrobial therapy (Figure 1). This is done by identifying which pathogens are present in the sample by culturing them. These pathogens can then additionally be tested for antimicrobial resistance (
-
What factors can prompt the sending of samples?
-
Factors that trigger a request for microbiological tests on a sample include the following:
- The physician wants to determine if infection is present.
- Standard antibiotics are not working (treatment failure).
- A very severe or unusual infection.
- Local policies or guidelines on sampling.
- Financial or other external incentives. For example, if a contract means the physician or healthcare organisation gets paid more if extra samples are sent, more are likely to be analysed, some perhaps unnecessarily, leading to a waste of resources. Ideally, the decision to send a sample is made without financial considerations in mind.
- Local culture or peer pressure, since people are likely to send, or not send, samples for testing according to what their colleagues are doing.
-
What factors can deter the sending of samples, even if clinically necessary?
-
Factors that can deter physicians from sending clinical samples for testing include:
- difficulty in accessing a laboratory, perhaps due to logistical or transport problems
- a lack of equipment to obtain samples, for example sterile swabs or blood culture bottles
- a lack of trust in laboratory test results
- turnaround times which are too long to be useful
- cost pressures, for example the patient cannot afford the test or there is only a small or no budget available for testing
- no local culture of doing tests
- not understanding the benefits of testing.
Clinical specimens are also routinely used as a source of data for AMR surveillance. However, a significant proportion of patients who could have a microbiology test performed never get sampled and so no results are available for them. This introduces an element of bias into the data. As a consequence, more severe cases and treatment failures could be over-represented. Tertiary care cases could also have more samples sent than those in more remote settings which could affect the overall results if resistance patterns differ between the settings. For long term, sustainable AMR surveillance, however, it is much cheaper and simpler to use clinical samples and the associated data than establishing a study to sample more randomly. The data are also always available whereas a surveillance study might be a one-off event.
Whilst it is not possible to avoid all potential bias when using clinical samples for surveillance purposes, it helps to be aware that reported rates of AMR may not be representative of the country or region as a whole.
1.2 Impact of sample quality on testing and reporting of results
Not all samples received by a laboratory will be of high quality, that is, contaminant free and in sufficient quantity. Prior to starting the testing process, laboratory staff should be aware of potential quality issues relating to specific sample types that could lead to false results.
Specimen contamination
It is important to recognise that when samples are taken from most sites in the body, the environment is not sterile. This means that a mix of the following may be present:
- Normal
commensal flora : Bacteria (or other microorganisms) that are usually found at that location in or on the body, e.g. skin, genital region, mouth, etc. For example,normal flora are likely to be found on superficial swabs, genital swabs and in respiratory specimens. Colonising organisms : Potentially pathogenic organisms not usually found at that part of the body, and may be displacing the commensal bacteria.- Contaminants: Microorganisms collected during specimen collection, e.g. from the patient, healthcare provider taking the sample or on the collection tube.
Blood and
Reducing contamination
Clinical staff can take several measures to improve the quality of the specimens they send to laboratories, either by reducing contamination through aseptic techniques and/or increasing the yield of true pathogens. For example, sterile containers and equipment should be used for sample collection, to avoid the introduction of contaminating organisms. Specimens need to be large enough to handle effectively and to minimise changes such as pH, drying out, etc., that may affect the viability of the organism concerned.
Some types of sample may require special treatment or have other considerations.
- Sputum samples, for instance are very useful for diagnosing
TB and are frequently sent to diagnose other respiratory tract infections. However, the results have to be considered carefully as the sample is likely to be contaminated with the commensal flora in the upper respiratory tract. Identifying all the organisms in a sputum sample will not help the diagnosis and takes up a lot of laboratory time and resources. - Blood cultures, which are important in diagnosing severe and/or bloodstream infections, are another sample type that is easily contaminated by the commensal flora of the skin. Even though the blood is taken carefully to minimise contamination and inoculated directly into blood culture media by the person taking the samples, contaminants will still sometimes be found. A video showing safe and accurate blood culture collection using ANTT (The Hillingdon Hospitals NHS Foundation Trust, 2019), shows how careful decontamination of the skin and bottles, plus aseptic technique can minimise sample contamination. Video 1 illustrates some of the key features of aseptic technique.
- Urine samples need to be collected carefully in midstream, to help reduce contamination with the commensal flora in the genital region. The sample should be refrigerated if it cannot be processed immediately. Boric acid can be used as a preservative if this is not possible. Microscopy helps determine whether a urinary tract infection (
UTI ) is present before culturing by looking for pus cells which indicate inflammation/infection.
Transcript: Video 1 Aseptic technique
Microbes are grown in the laboratory as cultures. Cultures can be grown from samples from a variety of sources including clinical specimens. In their natural environments, many species of bacteria grow together, but if we want to study a particular species, we need to grow it on its own in a pure culture. In a pure culture, all the cells present are of the same type. Pure cultures are maintained by sterilising media before use and by a practice called ‘aseptic technique’.
Aseptic technique prevents other microbes contaminating our culture and also prevents cultured microbes from escaping into the environment, which is very important when we’re dealing with pathogens.
To grow microbes, we must supply all their nutritional requirements by growing them in culture media. Media can be liquid, when they are called broths, or they can be solidified with a jelly-like substance derived from seaweed, called agar.
A wide range of different media are available as different bacteria have different nutritional requirements. In fact microbes can sometimes be identified by the media they can or cannot grow on.
Media must be sterilised before use and this is normally done in an autoclave, a type of industrial pressure cooker. Materials are sterilised by exposure to high pressure steam at a temperature of over 121 °C for 15 minutes. Sterilising media before use means that the only cultures that grow are the ones we put there.
To check whether sterility (the complete absence of living organisms) has been achieved, the materials are marked with special tape that changes colour under these conditions.
To prevent the spread of any pathogens into the environment, bacterial cultures that are not required any more are sterilised by autoclaving.
Laboratory staff also need to take care not to introduce contaminants into cultures. Good laboratory practice should be followed at all times.
Reporting results
After testing it is important to avoid reporting any additional organisms and instead focus only on the causative pathogen, if found. Correct reporting is needed to guide accurate treatment decisions and for surveillance purposes.
-
What might be the consequence if organisms of the commensal flora are reported in test results?
-
The reporting of commensal organisms in microbiology tests can lead to unnecessary or inappropriate antimicrobial treatment. For surveillance, it could make the results inaccurate.
1.3 Transport, labelling and processing of samples
Specimens should be sent to the laboratory as quickly as possible so that the causative agent is still viable (able to replicate and divide) when it gets there, and is not overwhelmed by the growth of commensal organisms or contaminants. A variety of techniques are used to achieve this aim and to minimise the effects of any delay. Refrigeration and carefully designed transport media can keep the organisms viable for longer but add to costs.
Each sample should be uniquely labelled and registered in a database so that it can be attributed to a specific patient, and its analysis prioritised according to the urgency with which the result is required. It is important to make sure results are stored securely to protect the patient’s confidentiality.
Activity 3: Processing specimens in your workplace
Use the question prompts below to think how clinical specimens arriving in your laboratory are processed.
- How are samples recorded? Do you have an electronic system or database or are samples recorded in a book? What steps do you take to protect patient confidentiality?
- Is there anything that can be improved in your system?
Discussion
Things to consider are listed below; you may have thought of other points.
Patient confidentiality is important so that personal information in relation to the diagnosis including microbiology requests and results is kept safe – whether this is electronic or paper based. If the laboratory is small, it may be safer and easier to write the results in a book rather than setting up a database. For large numbers of specimens an electronic system is faster, but data must be kept secure and with safeguards in place to prevent data loss if the system crashes.
It is important to understand how the physicians will be using the results and work with them to make sure what you are giving them is useful to them. A close working relationship also helps them understand what the laboratory can and cannot do. Physicians need results as soon as possible to guide treatment. It is useful to monitor ‘turnaround times’ – how long it takes between receiving the specimen and issuing the results. Sometimes issuing interim results, such as the Gram stain result, may also be helpful.
Not all testing of isolates can be carried out in a hospital laboratory and specimens/cultures may need to be sent to a reference laboratory for further processing. For example, some organisms may be difficult to culture or may need more advanced techniques to identify to species level (see Section 3.3).
Activity 4: Preparing samples for transfer to a reference laboratory
What factors do you need to consider when sending isolates to a reference laboratory?
Discussion
Factors you may have considered are listed below; you may have thought of others.
- The basic growth requirements for most pathogens are catered for by culture on a nutrient, blood or a chocolate agar slope and stored at room temperature.
Fastidious organisms have specific requirements and may also need special transport conditions otherwise they may not survive the journey. It may not always be possible to send these.- Some isolates may require refrigeration or freezing, this may not be practical in your laboratory. Refrigerated transport may be required for such samples.
- If freezing of cultures is essential then it must be done properly, for example with beads and glycerol, so as not to kill the organisms. Once frozen, cultures should not be thawed until use as repeated freeze-thaw cycles lead to deterioration.
- Safe transport must be arranged for hazardous organisms or samples.
- It is important to consider how long cultures remain viable. It may be necessary to keep some in reserve and send batches for analysis.
Accurate labelling that ensures patient confidentiality is important. Generally, isolates are given a number that can then be cross-referenced back for full identification.
2 Basic tests used to isolate and identify bacterial pathogens
We turn now to a description of the laboratory tests used to identify pathogens.
2.1 Culture media
Growing isolates from clinical samples in the laboratory is a useful part of diagnosing the cause of an infectious disease and is an essential step for determining the antibiotic sensitivities of bacteria.
All artificial media comprise a mineral base, a nitrogen source, a carbon source and an energy source. Supplements of organic compounds may be added to encourage or suppress the growth requirements of particular microorganisms. Media may be in liquid (broth) or solid form.
Most clinical samples are inoculated directly onto solid, agar-based culture media on a plate. Videos 2 and 3 demonstrate two common methods for preparing plates, streak plating and spread plating.
Transcript: Video 2 Streak plating
All bacteria in a single colony are usually identical. In a pure culture all the colonies will be alike in shape, size, colour, texture and smell. But in a mixed culture, like a clinical specimen, several different types of colony may be seen.
To check whether a culture is pure we use a technique called streak plating.
The first step is to label the plate with the date and the identity of the sample.
Labels are written on the part of the plate containing the agar so that if the lid is lost the culture can still be identified.
The Petri dish is kept upside down to minimise the chances of a contaminant falling onto the agar.
To make a streak plate we use aseptic technique and a bacteriologist’s loop.
A bottle containing the specimen is taken, flamed to remove any contaminating organisms, and the loop used to sample the microbes within. The loop full of microbes is touched to the surface of the agar and is spread across the plate in a series of streaks that act to successively dilute out the sample.
The plate is turned between each set of streaks. The final streak, a wiggly line, should allow individual microbes to be separated.
In order to more clearly see the streaking technique, we’ve drawn the pattern of streaks on the bottom of a Petri dish, so you can see the order in which they are made.
Bacteria that can grow in the human body are incubated at 37 °C.
After overnight incubation, at the more dilute end of the streak series, separate colonies, each resulting from the growth of a single microbe can be seen.
Transcript: Video 3 Spread plating
Using aseptic technique, we take out 0.1 ml of a particular dilution from its tube and place it onto the middle of the agar surface.
The inoculum is spread over the plate using a spreader. This one is pre-packed and sterilised. The spreader is moved over the surface of the agar, while the plate itself is rotated with the other hand to give an even spread of bacteria across the surface.
When grown at optimal temperatures in suitable conditions, this process enables the laboratory technologist to view individual colonies and select them for additional work according to their appearance (colonial morphology) and relative abundance. Table 1 provides details of the main media types used routinely in microbiology laboratories. Note that media may fall into more than one of the categories listed, for example it may be both selective and indicator.
| Media | Use |
|---|---|
| Basic | Standard nutritional content for non-fastidious organisms. Also used as a base for other media. |
| Selective | Contains substances (antibiotics, bile salts etc.) which suppress certain organisms. Used for specimens where a lot of competing |
| Enriched | Contains extra nutrients (blood, vitamins etc.) for |
| Indicator | Contains substances which change colour, for example when carbohydrate fermentation leads to acid production by certain organisms. |
| Chromogenic | Contains chromogenic substrates that change colour in response to the presence of bacterial enzymes. The colour change is species specific, therefore organisms can be identified by colony colour. Allows rapid, basic identification but is relatively expensive and requires well-controlled storage conditions. |
Activity 5: Culture media in your workplace
Use Table 2 to list the types of agar-based media you have in your laboratory and what you use them for. Note which category of media described in Table 1 each comes into. What considerations govern your laboratory’s choice of media?
| Name of medium | Use | Category |
|---|---|---|
Discussion
You may use many of the media in the example provided or use others not on the list. The media available to your laboratory will depend on factors such as local suppliers, transport and logistics, preferences in your country/region, and cost.
There may be more up-to-date media than the ones you are using as the technology continues to advance. These media save a lot of time, but may be too costly or complicated to use for many district laboratories or for those processing small numbers of specimens. Ready-made media save time in pouring, sterilising, and QC, but are not always available or practical outside of big cities.
| Name of agar medium | Use | Category |
|---|---|---|
| Nutrient | General purpose; supports growth of a wide range of non-fastidious organisms | Basic |
| Blood | For |
Enriched |
| Chocolate | Growing the most fastidious organisms | Enriched |
| CHROMagarTM Orientation urines | Identify common pathogens quickly | Chromogenic |
| Urine samples: only culture those samples with positive microscopy. This medium has characteristic colony appearance and colour for the common pathogens. It also stops Proteus from swarming over the plate. | Selective Indicator | |
| MacConkey | Isolate Gram-negatives | Selective Indicator |
| Modified New York City ( |
Isolate Neisseria gonorrhoeae | Selective Enriched |
| Mueller-Hinton ( |
Enriched | |
| Xylose Lysine Deoxycholate ( |
Isolate Salmonella and Shigella species | Selective Indicator |
2.2 Processing blood cultures
Blood cultures are inoculated directly from the patient into sterile liquid media. These media may be locally made, or commercial culture bottles can be purchased. The bottles are incubated in the laboratory, observed and sub-cultured. They are plated out onto agar plates for further work to determine if the blood culture is positive. Sub-culturing on a mix of media types is necessary to isolate most of the organisms likely to cause bloodstream infections.
-
Which types of organisms would you expect to isolate when sub-culturing onto blood agar in atmospheric oxygen, MacConkey in atmospheric oxygen, and chocolate agar in CO2.
-
With these culture media and conditions you should be able to isolate the main pathogens causing bloodstream infection. The chocolate agar and the CO2 will allow
fastidious organisms like Streptococcus pneumoniae and Neisseria species to grow. Most other organisms will grow well on blood agar in atmospheric conditions. Colonies can be taken from this for the next stages of testing. MacConkey is used to help in the initial identification of Gram-negative organisms (see Section 4.3).
There are also a number of automated systems in common use, such as BD BACTECTM, or BacT/ALERT® (Biomerieux) which use pre-prepared bottles (Figure 2). These can monitor the samples for growth and indicate when it is time to do the sub-culture. Although the equipment is expensive, the risk of contaminants being introduced is reduced and turnaround times are usually faster too, as the subculture can be set up as soon as the equipment indicates positive growth, instead of once daily as is usual for manual blood cultures.

2.3 Gram stain
The Gram stain test allows you to see basic bacterial morphology and group bacteria into two main types; Gram-positive and Gram-negative.
Watch Video 4 to see how the test is performed. If you are unable to watch this video, you can view a transcript of the content by clicking on ‘Show transcript’ below.

Transcript: Video 4 How to perform a Gram stain
The Gram stain is the most important stain in microbiology. The first step is to heat-fix the bacteria to a microscope slide. Using a sterile loop, a few drops of water are transferred to the slide. The culture is then mixed with the water to give an emulsion and this is spread onto the slide. The secret of a good Gram stain is not to make the emulsion too thick. The emulsion is left to dry at room temperature.
Then the slide is passed through the Bunsen flame to fix the bacteria to the glass, so that they won’t be washed off by the staining. It’s important not to let the slide get too hot or the bacteria will burst. If it’s too hot for you, it will be too hot for the bacteria.
In Gram’s staining, the slide is flooded with crystal violet stain and left to stand for one minute. The crystal violet is washed off with slow-running tap water. The next step is to flood the slide with Gram’s iodine, which is also left for a minute. The iodine forms a complex with the crystal violet, giving a dark purple colour. The slide is rinsed again before the next step.
Next, 95% alcohol is used to wash the purple dye away. Some bacteria retain the dye; others don’t. After a rinse in water, the final step is to add a red dye, Safranin, which is left on for just 30 seconds.
After a final wash, the slide is blotted, dried, and then examined under the microscope. Some bacteria appear pink. Since they lost the original purple colour, they are called Gram negative. Other bacteria retain the purple colour, which masks the red dye, and these ones are called Gram positive. Round bacteria are called cocci. Rod-shaped bacteria are often known as bacilli.
Note that laboratories without a gas supply can still perform Gram stain tests by using single use sterile plastic loops which are disposed of into disinfectant and the slide may be fixed by flooding with methanol (a technique that is preferred in some cases anyway).
Activity 6: Troubleshooting
The Gram stain may seem like a simple test, but things can still go wrong. Think about your own laboratory practice – why might Gram-positive organisms appear Gram-negative? What measures can you take to prevent this happening?
Discussion
Factors that can lead to incorrect results for the Gram stain include:
- excessive heat fixation damaging the cell wall
- over-decolourisation, so that the stain is washed out
- old or badly stored iodine
- using an old culture to do the smear – organisms are damaged.
Measures that can be taken to prevent errors include:
- following laboratory
SOP s exactly - making sure reagents are dated and renewed regularly – if reagents are stored long enough bacteria can grow in them and appear on the slides!
- only using fresh cultures.
3 Commercially available and more advanced tests
Once the specimens have been cultured and single colonies are available growing on a plate, additional testing is done to determine the bacterial species. Many laboratories rely on standard biochemical testing. Biochemical test strips are available which enable multiple tests to be performed at the same time.
3.1 Biochemical test strips
Multiple biochemical tests in a single kit form, come as a strip with micro wells and reagent. The testing process is straightforward; inoculate, incubate, add some more reagents then read and compare the result to a standard. Examples of testing kits include Analytical Profile Index (
Activity 7: API identification of Gram-negative pathogens
In this activity you will first watch a video showing how to set up an API test for the identification of Gram-negative pathogens, and then answer some related questions.
First, watch the video ‘Performing an API20E strip test’ (2016).
Note that in the video Enterbacterales are referred to by the old term ‘Enterobacteriaceae’.
Transcript
Add water to 3/4 fill all of the honeycomb wells of the tray
Flame a loop until it is red hot then allow it to cool for 20 seconds
Select a single isolated colony and emulsify in 10 mils of sterile distilled water
Use a new sterile pipette
For CIT, VP and gel tests fill both the tube and cupule
Fill only the tubes for the remaining tests
Add one or two drops of paraffin oil two tests: ADH, LDC, ODC, H2S and URE
Incubate the strip at 37°C overnight
After incubation add biochemical reagents to the required tests
One drop each of VP1 and VP2 to the VP test
One drop of Kovacs reagent to the IND test and
One drop of the TDA reagent to the TDA test
After 10 minutes compare the colours against the chart and record on an API record sheet
Now, answer the following questions:
- What temperature is the test kit incubated at overnight?
- Would the same API kit be suitable for all of the following: E. coli, Shigella, Salmonella, Klebsiella, Haemophilus influenzae?
Discussion
- The test kit should be incubated at 37°C overnight.
- No. ‘E’ in API20E stands for Enterobacterales (formerly known as Enterobacteriaceae), so all of these organisms are identified by API 20E except Haemophilus influenzae. Haemophilus influenzae, as a non-glucose fermenter, is not classified as Enterobacterales. A different kit (API 20NE – ‘Non-Enterobacterales’) would be needed to identify Haemophilus influenzae.
Activity 8: Individual biochemical tests versus test kits
What are the pros and cons of using individual biochemical tests rather than test kits such as API or similar?
Discussion
| Test type | Advantages | Disadvantages |
|---|---|---|
| Individual |
|
|
| Test strip kits |
|
|
3.2 Chromogenic media
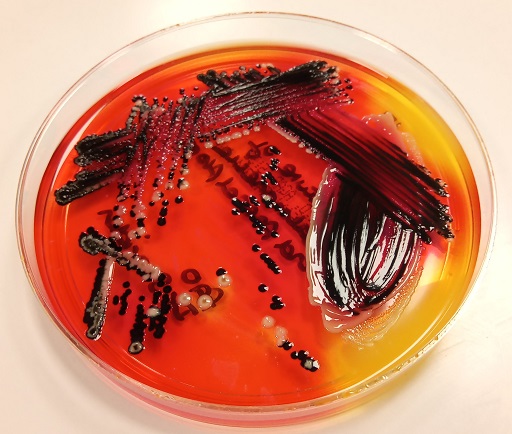
Chromogenic media, first mentioned in Section 2.1, are useful in certain situations, for example to identify:
- organisms in potentially mixed samples – if you have a lot of samples to test (see Figure 3)
- specific species with a reasonable level of accuracy without the need for additional testing
- resistant strains/species.
Whilst expensive, this relatively new media type saves a lot of time in laboratories that deal with large numbers of routine urine samples.
Activity 9: Species identification
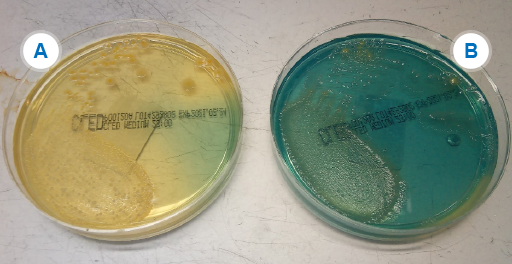
In Figure 3B, Klebsiella and Enterobacter look the same (metallic blue) and for clinical purposes are usually reported as ‘coliform species – non-E. coli’. However, for management and for surveillance purposes, it may be necessary to identify K. pneumoniae from other coliforms, particularly if there is concern about MDR strains of K. pneumoniae.
What options would you have to differentiate the Klebsiella species from other non-E. coli coliforms?
If you are not able to answer this question now, you can return to it after having studied other sections of this course.
Discussion
- API20E,
MALDI-TOF (see Section 3.3.1), and automated systems would all be able to differentiate these bacteria. However, if none of these is available, motility and biochemical tests are performed. Although these two organisms are very similar Enterobacter is motile whereas Klebsiella is not so the motility test is a useful way of discriminating between them. - The additional tests might be too time-consuming and/or expensive to do on every isolate just for surveillance. However, if the tests are not done then this could potentially be a source of bias and error in AMR data.
- One option is to send the isolate to a reference laboratory for testing, but if only the more resistant isolates are sent this would again be a potential source of bias and error in AMR data.
- A better option might be to implement a sampling framework where a proportion of isolates received by the laboratory are identified for AMR surveillance purposes.
3.3 Advanced testing
Some testing methods are beyond the scope or resources of the hospital laboratory and are more likely to be found in public health and reference laboratories.
3.3.1 MALDI-TOF
Matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) (Croxatto et al., 2012) gives a result faster than most standard biochemical test methods and at a lower cost per sample. It is both accurate and reliable, and gives identification to species level from a pure culture within minutes. In recent years, MALDI-TOF has revolutionised microbiology practice in laboratories where it is used, having largely replaced most biochemical identification methods.
Disadvantages of MALDI-TOF are that the equipment is expensive and often requires expensive maintenance contracts. It is also not effective for all organisms. For example, it cannot distinguish well between E. coli and Shigella, nor between S.pneumoniae and viridans type Streptococci. For these, additional biochemical testing is still needed for confirmation.
The rapid throughput means that hundreds of isolates can be processed daily using very little technologist time, meaning that it is most cost-effective for laboratories with large sample throughputs (identifying several thousand isolates per year). For these laboratories, the cost of the instrument and the maintenance contracts is very rapidly offset in savings on reagents and staff time.
It is possible to use MALDI-TOF to identify a small number of resistance mechanisms (e.g. for
If you are not familiar with MALDI-TOF, watch the following short video about this method (Theory of MALDI-TOF Mass Spectometry, 2016).
3.3.2 Automated systems
Automated systems, such as Vitek II (Biomerieux), BD Phoenix (Beckton Dickinson) and Beckman-Coulter-MicroScan are used in some clinical laboratories. Effectively these are an automated version of the biochemical test strips, with more reagents and analytic software. The cost of such systems is high, but less work is needed to complete the tests. A further advantage is that AST can be done at the same time.
3.3.3 Commercial PCR
Commercial
Two commercial PCR systems are:
- Cepheid’s GeneXpert® System, a family of workstations that can run 2–80 PCR tests on a single, consolidated workstation.
- The BioFire® FilmArray® System, a scalable system with multiplex PCR testing for infectious disease diagnostics.
3.3.4 Whole genome sequencing (WGS)
Whole genome sequencing (WGS) is a method for rapid sequencing of the chromosomal DNA and mitochondrial DNA (and chloroplasts DNA in plants). WGS is carried out using next-generation sequencing (NGS) technologies, which allow the rapid generation of large quantities of genomic data.
WGS has the potential to become an important tool in pathogen surveillance. However, the challenge in AMR is the timely processing of the large volumes of data, particularly during emergency pathogen outbreaks (Bogaerts, 2021 and Jauneikaite, 2023). As a result, the Surveillance and Epidemiology of Drug-resistant Infections Consortium (SEDRIC, n.d.) working group are working on a series of recommendations on the use of genomic surveillance via WGS for AMR. The working group has identified various advantages and barriers to the use of WGS in AMR:
Advantages
- Health-care associated infections in hospitalised patients pose a significant challenge globally. Genomic AMR surveillance of pathogens using WGS from health-care associated infections can provide much better identification of the causative agent, such as genomic subtypes. It can also help to identify facility-level trends.
- One particular advantage of WGS is its use to investigate outbreaks and to improve support for infection prevention and control in AMR surveillance in health-care facilities. It can help to identify complex epidemiological patterns, such as the emergence of new strains and expansion of new multidrug-resistant strains.
Barriers
- Despite many advantages, there are still many barriers to WGS in AMR surveillance. The initial set up and running costs can be prohibitive or the equipment difficult to obtain due to poor distribution networks and supply chains.
- There are also significant challenges in the analysis and interpretation of the genomic data, which typically requires bioinformaticians. Quality assurance processes for both laboratory sequencing and bioinformatic analysis have still yet to be clearly defined.
You will learn more about WGS in the Whole genome sequencing in AMR surveillance course.
4 Identifying the key pathogens in global AMR surveillance
4.1 The thirteen key pathogens
Thirteen key bacterial pathogens in humans have been identified by the World Health Organization (
| Pathogen | Gram reaction |
|---|---|
| Staphylococcus aureus | Gram positive |
| Streptococcus pneumoniae | |
| Acinetobacter species | Gram negative |
| Escherichia coli | |
| Haemophilus influenzae | |
| Klebsiella pneumoniae | |
| Neisseria gonorrhoeae | |
| Neisseria meningitidis | |
| Pseudomonas aeruginosa | |
| Salmonella species (non-typhoidal) | |
| Salmonella enterica serovar Typhi | |
| Salmonella enterica serovar Paratyphi A | |
| Shigella species |
All of the GLASS target pathogens are listed on the WHO’s priority pathogen list (WHO, 2024). E. coli, K. pneumoniae and Acinetobacter all have globally widespread strains which are multi-drug resistant (
-
Why has N. gonorrhoeae been chosen out of all the pathogens causing sexually transmitted infection (
STI ) as a focus for surveillance? -
Cephalosporin and/or fluorquinolone-resistant N. gonorrhoeae is listed as a high priority pathogen on the WHO’s priority pathogen list (WHO, 2024). Infection with N. gonorrhoeae leads to a lot of morbidity including pelvic inflammatory disease and infertility. Resistance is a big problem – especially to oral antibiotics. Some strains are already only sensitive to
IV antibiotics, and untreatable strains are a becoming a possibility. N. gonorrhoeae can be cultured in a clinical laboratory whereas it is not possible to test the organisms that cause Chlamydia, Syphilis etc. for AMR. Resistance is, fortunately, less of an issue so far with these organisms.
Other GLASS target pathogens affect vulnerable populations; for example, H. influenzae, which can cause severe infections in children.
Activity 10: Usual isolation site of GLASS target pathogens
For each of the images below, can you identify the site you would expect them to be isolated from? Note: for some pathogens, there may be more than one site.
Click 'reveal discussion' to see the answer.
Which site or sites in the body is S. aureus isolated from?
Discussion
S. aureus is a common cause of infection in the community and of healthcare associated infection (
For
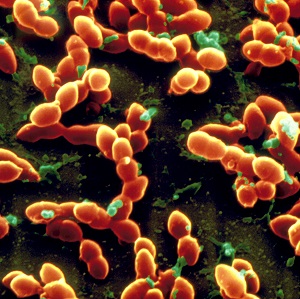
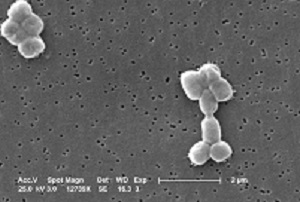
Which site or sites in the body is S. pneumoniae isolated from?
Discussion
S. pneumoniae is a common cause of pneumonia, meningitis and ear infections, as well as infections in HIV patients. It is typically isolated from bloodstream samples in severe infections. It can also be found in upper respiratory and surface swabs, sputum and
GLASS surveillance focuses on bloodstream, CSF and lower respiratory tract infection isolates.
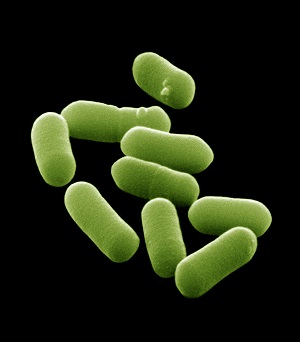
Acinetobacter are gram-negative, coccobacillary-shaped bacteria. A. baumannii accounts for most Acinetobacter infections in humans and is the species of concern. However, identification to species level is technically challenging so many sites report as Acinetobacter species only.
Which site or sites in the body are Acinetobacter species isolated from?
Discussion
Acinetobacter are classic
For GLASS, bloodstream and lower respiratory tract isolates are the focus of interest.
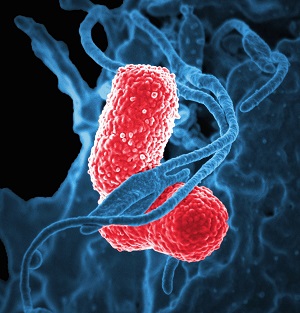
Which site or sites in the body is E. coli isolated from?
Discussion
E. coli is the most common cause of
E. coli is typically isolated from bloodstream and urine. They can also be isolated from stools, but this requires special techniques to distinguish the pathogenic strains from commensal E. coli which is prevalent in the gut.
For GLASS, isolates from bloodstream and urine are the focus of interest.
Which site or sites in the body is K. pneumoniae isolated from?
Discussion
K. pneumoniae is a common cause of HCAI including
K. pneumoniae is typically isolated from bloodstream and urine samples and GLASS focuses on these as well as lower respiratory tract specimens. It can also be found in swabs and sputum samples, but it is harder to be sure it is acting as a pathogen rather than colonising these sites.
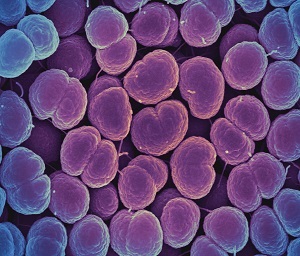
Which site or sites in the body is N. gonorrhoeae isolated from?
Discussion
N. gonorrhoeae causes the
For GLASS, N. gonorrhoeae isolated from genital samples are the focus.
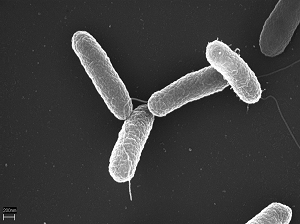
Which site or sites in the body are Salmonella species isolated from?
Discussion
Enteric fever is caused by S. typhi/paratyphi with the other non-typhoidal strains causing diarrhoeal disease. Enteric fever strains only affect humans, but the other strains can also be transmitted via animals and animal products. This is important in the
For GLASS, Salmonella spp. (non-typhoidal) isolated from both stool and blood samples are the focus. For Salmonella enterica serovars blood samples are the focus.
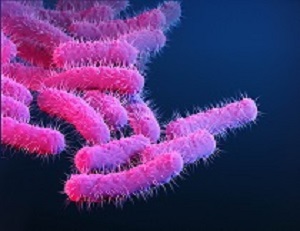
Which site or sites in the body are Shigella species isolated from?
Discussion
Shigella species cause diarrhoeal disease and cause significant morbidity in young children.
For GLASS, Shigella isolated from stool samples are the focus.
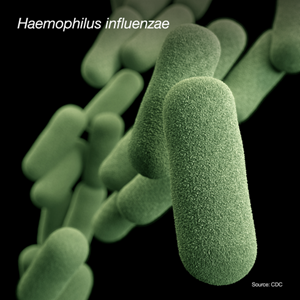
Which site or sites in the body is H. influenzae isolated from?
Discussion
H. influenzae can cause many different types of infections. These infections range from mild (ear infections in children) to serious (bloodstream infections). The most common serious infections caused by H. influenzae are pneumonia, bloodstream infections and meningitis.
For GLASS, H. influenzae isolated from CSF and the lower respiratory tract are the focus.
Which site or sites in the body is Neisseria meningitidis isolated from?
Discussion
Neisseria meningitidis cause meningococcal disease. The two most common types of meningococcal infections are meningitis and bloodstream infections.
For GLASS, Neisseria meningitidis isolated from blood and CSF are the focus.
Which site or sites in the body is P. aeurginosa isolated from?
Discussion
P. aeruginosa can cause infections in the blood, lungs, urinary tract or other parts of the body after surgery. Patients in healthcare settings are at highest risk.
For GLASS, P. aeurginosa isolated from blood and the lower respiratory tract are the focus.
Your laboratory is almost certainly identifying other organisms than the ones listed in Activity 10. However, the focus of this course is on some of the issues around identifying the eight key GLASS pathogens under global AMR surveillance. Your laboratory should have comprehensive standard operating procedures (
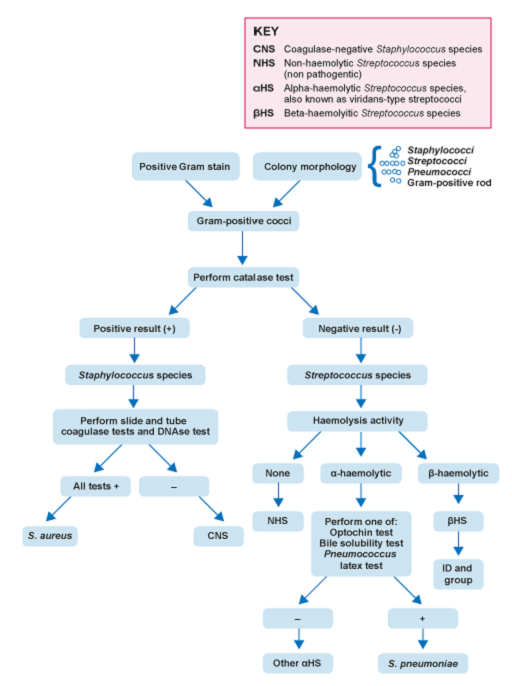
4.2 Identifying Gram-positive cocci
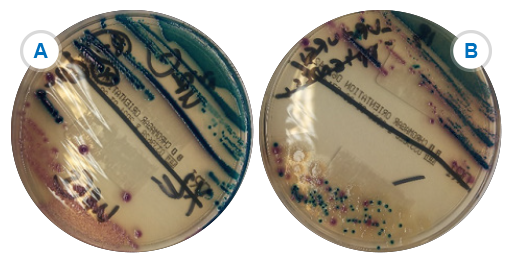
Your laboratory needs to be able to reliably confirm the identification of S. aureus and S. pneumoniae (see Figure 4).
Enterococci can show any of the three patterns of haemolysis. They are generally Lancefield Group D can be confirmed using biochemical tests such as aesculin hydrolysis or litmus milk reduction. They ferment a range of sugars including lactose.
For S. aureus, a presumptive identification is done by the characteristic appearance of colonies on agar and a positive catalase test. A minimum of two tests from the following must be used to confirm identification: tube coagulase, slide coagulase, or DNAse. Alternatively,
-
Why is more than one test needed to confirm S. aureus?
-
Bacteria, being living organisms, will occasionally not test the way the book says they should, so more than one test is needed to confirm. Some ‘coagulase negative’ Staphylococci will test positive on one of the coagulase tests. This is important for clinical reasons, to give the right treatment, and also for
AMR surveillance. For example, methicillin-resistant, coagulase-negative Staphylococcus could be mis-reported as MRSA leading to inappropriate and unnecessary antibiotics and providing inaccurate surveillance data.
-
What system of controls for tests should be in place?
-
For any identification, not just for S. aureus, it is important to use positive and negative control organisms to make sure your reagents are working correctly. Ideally these organisms are reference strains obtained from the UK National Collection of Type Cultures (
NCTC ) or the American Type Culture Collection (ATCC ). For these tests the controls are set up in parallel with the test organism.
S. pneumoniae is
Activity 11: Testing for S. pneumoniae in your workplace
Think about your workplace activities and consider these questions.
- If you are able to culture and identify S. pneumoniae reliably in your laboratory, how are you differentiating it from other streptococcal species?
- If isolation of this organism is not reliable, or you do not usually work with this organism, what might be practical barriers to its identification?
- How might laboratory practice, for example the ease of isolating S. pneumoniae, affect AMR surveillance for this organism?
Discussion
- Differentiation from other streptococcal species can be by appearance, haemolysis, specific confirmatory tests and by the fact that it only grows on blood-containing media under CO2.
- CO2 for incubation, and blood-containing media are both necessary for growth so these are limiting factors. Autolysis can occur in blood cultures. This is when the organism grows so fast in some media that it outstrips its nutrient supply, and no viable organism is detected on subculture. So you will need to have access to the right culture media and conditions to isolate this organism.
- Under-reporting from some laboratories who struggle to isolate the organism versus better equipped laboratories, could lead to bias if antibiotic resistance rates varied between the settings.
4.3 Identifying Gram-negative organisms
Gram-negatives are found in a range of clinical samples. As they are common causes of
The approach taken to identify Gram-negative organisms depends on several factors.
- The site of the body where the sample was taken. In Activity 10, for example, you learned which type of sample would be most likely to yield which key pathogen.
- The likelihood of the sample from this site containing a mixture of organisms – commensals or contaminants
- Whether there is already a presumptive identification. For example, stool samples are usually cultured on media which selects for enteric pathogens. When S. typhi and S. paratyphi are suspected, blood cultures may also be taken to look for these organisms. The identification process of these organisms from blood cultures will be longer than from stool samples as you do not have the benefit of seeing the appearance on the selective medium.
In practice, identification of Gram-negative organisms often proceeds by a process of elimination; one positive test result dictates the choice of a secondary test and so on. This process requires laboratory technologists to have the appropriate knowledge and skills to perform this screening process.
The following sections outline how the six Gram-negative key
4.3.1 Enterobacterales species
E. coli and K. pneumoniae are two amongst a variety of coliforms (Enterobacterales).
Your laboratory is likely to be using a differential, indicator medium based on lactose fermentation – usually either
Activity 12: Identifying Enterobacterales in your laboratory
How do you identify the different Enterobacterales species (also known as Enterobacteriaceae or coliforms) in your laboratory? Make some notes and then compare with the sample answer.
Discussion
There are a number of ways of identifying Enterobacterales species depending on budget, number of specimens handled, and relative cost of staffing time versus reagents, available equipment etc.
Methods you may have listed include:
- Indicator media.
- Motility test, biochemical tests (for example, indole test, Voges–Proskauer test etc.)
- Commercial options such as disk forms of the biochemical tests, for example Rosco DiatabsTM or biochemical strip tests, for example
API or Microbact™ - Automated systems or
MALDI-TOF .
The UK Standards for Microbiology Investigations (
-
Why is it useful to identify Enterobacterales to species level for GLASS?
-
In many cases, species level identification is needed to properly apply the breakpoint tables and associated rules. It can also make it easier to spot when an organism has deviated from expected patterns of behaviour or developed mechanisms of resistance. For example, a plasmid may have jumped species or the organism acquired new resistance genes. It then becomes possible to focus on tracking and controlling the organism. On a local scale you can identify and control outbreaks quicker.
Resistant organisms of some species are more important pathogens from the Public Health point of view than others. For example, K. pneumoniae can cause big
K. pneumoniae is the only Klebsiella species which is a current focus for
As discussed in Section 3.2 though, only sending the resistant ones to the reference laboratory for species identification could lead to bias in the surveillance figures. This would make it hard to tell the prevalence of the resistant strains in the area accurately. However, it has the major benefit of allowing you to be aware of which resistant Klebsiella species and strains are present in your hospital.
4.3.2 Haemophilus influenzae
Identification of Haemophilus species requires a combination of methods. Colonies on blood or chocolate agar may be presumptively identified by colony morphology. Haemophilus species produce colonies that are flat, convex and grey-white on blood agar. Following presumptive identification, further techniques, including MALDI-TOF or biochemical tests can be used to further identify the species.
4.3.3 Enteric pathogens
Your laboratory will almost certainly identify a wide variety of enteric pathogens from stool samples. For GLASS, the focus is on Salmonella and Shigella.
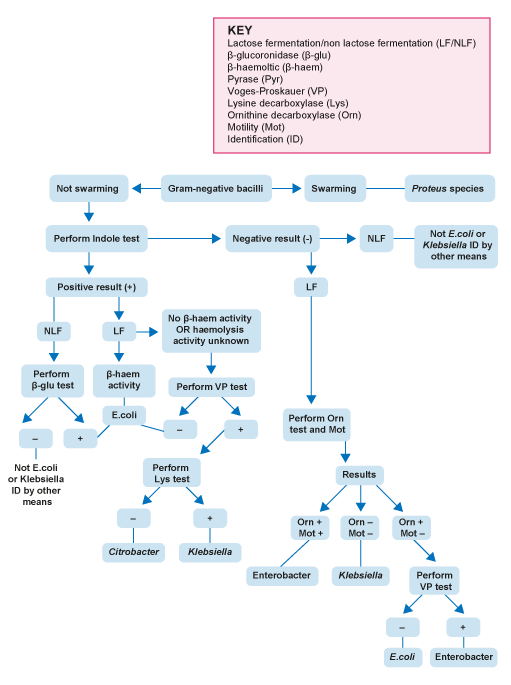
The first step in identification is the appearance of colonies on solid media. For example, on
-
Why is it important to do a urease test?
-
NLF enteric organisms, such as Proteus and related species (Providentia, Serratia and Morganella) have a similar appearance on solid media. It is therefore important to do a urease test to prevent mis-identifying these as enteric pathogens.
Confirmatory tests for Salmonella and Shigella include:
- a negative oxidase test – to differentiate from other NLF organisms like Pseudomonas
- Kligler’s Iron Agar (
KIA ) test - biochemical tests
- antisera agglutination tests
For details of all tests, see the UK
SMI guidance for Salmonella (PHE, 2021) and Shigella (PHE, 2022)
4.3.4 Pseudomonas aeruginosa
P. aeruginosa is a glucose non-fermenting Gram-negative rod bacteria that can grow under a variety of conditions, surviving temperatures of up to 42°C.
Pseudomonas species have no specific nutritional requirements and are non-fastidious; therefore, they grow well on all standard laboratory media. They produce colonies that are usually large and smooth with flat edges. P. aeruginosa is oxidase-positive; therefore, when coupled with colonial morphology, the results of the oxidase test can aid in providing preliminary identification for isolates.
4.3.5 Acinetobacter
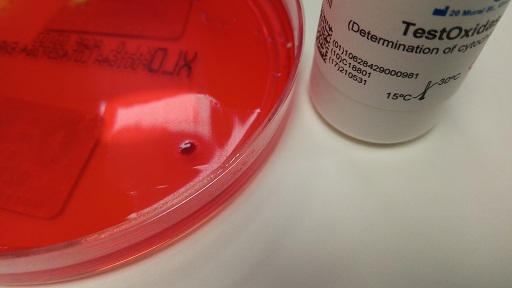
Acinetobacter are non-fermenting organisms which can be differentiated from other non-fermenters such as Pseudomonas species because they are oxidase negative (Figure 9). Biochemical tests like
4.3.6 Neisseria gonorrhoeae
N. gonorrhoeae is a
Initial identification from culture media is by Gram stain (Gram-negative cocci) and oxidase test (positive). Confirmatory tests include:
- individual biochemical tests or test strips (e.g. API-NH, Biomerieux)
- immunological tests, for example a slide-based co-agglutination test of which there are many commercial kits available (e.g. Phadebact® Monoclonal GC Test, MKL diagnostics).
-
Why is it necessary to confirm identification of N. gonorrhoeae with more than one test?
-
A lot of commensal Neisseria species can be found in the genital tract and it is important to distinguish between these and the pathogenic N. gonorrhoeae. Getting this wrong can have negative repercussions for the patient and their relationships as well as for AMR surveillance. This particularly applies if the patient is not known to be at high risk of
STI .
4.3.7 Neisseria meningitidis
Neisseria meningitidis is closely related to Neisseria gonorrhoeae but causes an entirely different disease. Like N. gonorrhoeae it is fastidious, requiring similar specialist media and growth conditions. Initial identification from culture media is by Gram stain (Gram-negative cocci) and oxidase test (positive) with follow up confirmatory biochemical, immunological and enzyme tests where MALDI-TOF is not available. At least two of these tests are required to confirm Neisseria species.
5 Fundamentals of quality control for isolating and identifying bacteria
In a clinical microbiology laboratory several factors can affect the accuracy and reliability of test results. Quality control (QC) measures are therefore put in place to monitor whether tests perform as expected and do so reliably. You will learn more about QC and other
Activity 13: QC in clinical microbiology laboratories
Think about what you have learned in this course. Why is QC important when identifying pathogens from clinical samples?
Discussion
QC ensures the following:
- You can actually grow the organisms you’re looking for.
- Selective media are inhibiting the appropriate organisms and allowing the growth of others.
- You are identifying organisms correctly.
- Your media and test reagents are working correctly, for example if they have deteriorated during storage or there is something wrong with what the manufacturer has supplied, the QC tests will pick this up.
- You are performing the tests under optimal conditions, for example the incubator is at the right temperature.
- Organisms are correctly identified before
AST , which is species-specific, is performed. - The results from the laboratory can be relied on as accurate for both clinical and surveillance use.
- Ultimately patients get better treatment as a result of timely and accurate diagnosis.
6 Identifying pathogens in your workplace
You should now have a better understanding of the microbiology techniques used in a variety of settings for identifying bacteria, and in particular the
Activity 14: Identifying pathogens in your workplace
Think about the eight GLASS priority pathogens and then reflect and make notes in response to the following questions.
- Are you identifying all of these pathogens to the required level in your laboratory?
- Are you confident that the identification is accurate?
- Are there areas where you feel your laboratory may not be reliably identifying pathogens? Which organisms does this apply to?
- How could the diagnostic capacity of your laboratory be improved and what are the barriers to achieving this? For example, what extra equipment and reagents might you need? Are the correct controls being used? Do staff need more training?
Finally, think about how you would make a case to your Laboratory Manager to convince them to support you in improving your laboratory’s diagnostic capacity.
Discussion
You may find it helpful to discuss your thoughts with colleagues before talking to your Laboratory Manager.
7 End-of-course quiz
Well done – you have reached the end of this course and can now do the quiz to test your learning.
This quiz is an opportunity for you to reflect on what you have learned rather than a test, and you can revisit it as many times as you like.
Open the quiz in a new tab or window by holding down ‘Ctrl’ (or ‘Cmd’ on a Mac) when you click on the link.
8 Summary
In this course you have learned about microbiology tests used to isolate and identify key human pathogens which are the focus of the
You should now be able to:
- know where samples are obtained from and reflect on the process by which bacterial samples are processed in your workplace
- describe the principles of laboratory tests used to isolate and identify key bacterial pathogens in human health, that are the focus of the GLASS programme
- know when and why advanced testing such as mass spectrometry and automated systems are used
- know the importance of procedures designed to ensure the quality of laboratory work relating to isolating and identifying bacteria in your workplace.
Now that you have completed this course, consider the following questions:
- What is the single most important lesson that you have taken away from this course?
- How relevant is it to your work?
- Can you suggest ways in which this new knowledge can benefit your practice?
When you have reflected on these, go to your reflective blog and note down your thoughts.
Activity 15: Reflecting on your progress
Do you remember at the beginning of this course you were asked to take a moment to think about these learning outcomes and how confident you felt about your knowledge and skills in these areas?
Now that you have completed this course, take some time to reflect on your progress and use the interactive tool to rate your confidence in these areas using the following scale:
- 5 Very confident
- 4 Confident
- 3 Neither confident nor not confident
- 2 Not very confident
- 1 Not at all confident
Try to use the full range of ratings shown above to rate yourself.
When you have reflected on your answers and your progress on this course, go to your reflective blog and note down your thoughts.
9 Your experience of this course
You’ve now reached the end of this course. If you’ve enrolled on a pathway, please go back to the pathway page and tick the box to confirm that you’ve completed this course. On the pathway page you’ll see both your progress so far as well as the other courses you need to complete in order to achieve your Certificate of Completion for that pathway.
Now that you have completed this course, take a few moments to reflect on your experience of working through it. Please complete a survey to tell us about your reflections. Your responses will allow us to gauge how useful you have found this course and how effectively you have engaged with the content. We will also use your feedback on this pathway to better inform the design of future online experiences for our learners.
Many thanks for your help.
References
Acknowledgements
This free course was collaboratively written by Liz Sheridan and Sarah Palmer, and reviewed by Priya Khanna, Melanie Bannister-Tyrrell, Skye Badger, Claire Gordon, Natalie Moyen and Hilary MacQueen. The course was reviewed and updated by Ben Amos, Vikki Haley and Rachel McMullan in 2025.
Except for third party materials and otherwise stated (see terms and conditions), this content is made available under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 Licence.
The material acknowledged below is Proprietary and used under licence (not subject to Creative Commons Licence). Grateful acknowledgement is made to the following sources for permission to reproduce material in this free course:
Images
Course image: Yulia Koltyrina/123RF.
Figures 1, 2 and 4: L. Sheridan.
Figures 3, 5, 6, 8 and 9: E. Tinnion and L. Sheridan.
Activity 10 (top to bottom): S. aureus: National Institutes of Health/Stocktrek Images/Getty Images; S. pneumoniae: Eye of Science/Science Photo Library; A. baumannii: Janice Haney Carr (public domain image); E. coli: David McCarthy/Science Photo Library; K. pneumoniae: Cultura Creative RF/Alamy Stock Photo; N. gonorrhoeae: National Institute of Allergy and Infectious Diseases, National Institutes of Health, ‘Neisseria gonorrhoeae bacteria’, https://www.flickr.com/ photos/ nihgov/ 24503565430 – this file is licensed under a Creative Commons Attribution-NonCommercial 2.0 Generic (CC BY-NC 2.0) licence, https://creativecommons.org/ licenses/ by-nc/ 2.0/; S. typhimurium: Volker Brinkmann, Max Planck Institute for Infection Biology, Berlin, Germany in ‘A novel data-mining approach systematically links genes to traits’ (2005) PLOS Biology 3(5): e166, https://doi.org/ 10.1371/ journal.pbio.0030166 – this file is licensed under a Creative Commons Attribution 2.5 Generic (CC BY 2.5) licence, https://creativecommons.org/ licenses/ by/ 2.5/; Shigella: Stephanie Rossow/Science Photo Library/Getty Images; H. influenzae and Neisseria meningitidis: CDC/Sarah Bailey Cutchin; P. aeruginosa: CDC/Antibiotic Resistance Coordination and Strategy Unit.
Figure 7: adapted from Ng, L.S.Y., Thean Yen Tan, T.Y. and Yeow, S.C.S. (2010) ‘A cost-effective method for the presumptive identification of Enterobacteriaceae for diagnostic microbiology laboratories’ [online] DOI: https://doi.org/ 10.3109/ 00313021003631338
Tables
Table 3: based on Global Antimicrobial Resistance Surveillance System: Manual for Early Implementation, World Health Organization, surveillance methods, p. 5, copyright 2015, https://apps.who.int/ iris/ bitstream/ handle/ 10665/ 188783/ 9789241549400_eng.pdf .
Video
Video 4: The Open University.
Every effort has been made to contact copyright owners. If any have been inadvertently overlooked, the publishers will be pleased to make the necessary arrangements at the first opportunity.