2.1.2 Destroying or modifying the antibiotic molecule
The second mechanism of antibiotic resistance you will look at is the destruction or modification of the antibiotic by bacterial enzymes. Probably the most well studied example of enzymes that destroy antibiotics are the β-lactamases.
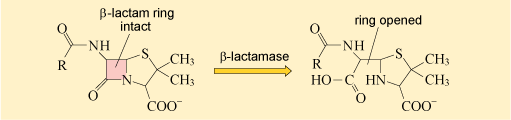
As you may recall, β-lactamases deactivate the β-lactam ring of β-lactam antibiotics, preventing them from binding to their target (Figure 17).
The first β-lactamase to be identified was penicillinase. As its name suggests, penicillinase can hydrolyse penicillin but not other, more recently discovered β-lactam antibiotics like cephalosporins. In the 1980s, a new group of β-lactamase enzymes were detected in Europe that hydrolyse a wider range of β-lactams. These enzymes are known as extended spectrum β-lactamase (ESBL).
ESBLs can deactivate almost all of the β-lactam antibiotics currently in therapeutic use. Consequently, their presence significantly reduces the available treatment options for infections caused by bacteria expressing β-lactamase. Since they were first described in the early 1980s, the frequency of infections caused by ESBL-producing bacteria has been increasing. Consequently, ESBLs represent an ever-growing healthcare challenge.
Some β-lactamases can be counteracted by using a β-lactamase inhibitor (a drug that can block the action of the enzyme, such as clavulanic acid or tazobactam) in combination with a β-lactam antibiotic (e.g. clavulanic acid and amoxicillin are combined in co-amoxiclav); however, even these are not necessarily effective against ESBLs, and each combination needs to be tested against the bacteria before using clinically.
How might a β-lactamase inhibitor help the treatment of infections caused by β-lactamase-expressing bacteria?
The β-lactamase inhibitor will block the ability of the β-lactamase to deactivate the β-lactam antibiotic so that the antibiotic can bind to its target molecule.
Other antibiotic-modifying enzymes do not destroy or target the core chemical structure that confers antibacterial activity. Instead they modify the antibiotic’s structure by adding chemical groups that prevent it from binding to its target. One group of antibiotics that are particularly susceptible to modification are the aminoglycoside antibiotics which include streptomycin and gentamicin (Figure 18).
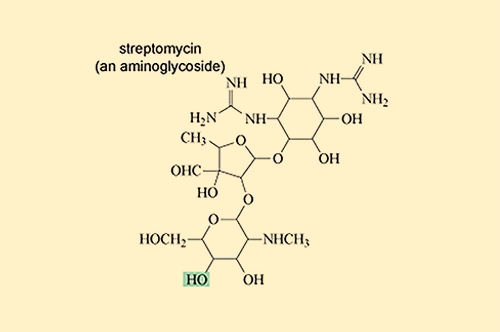
Aminoglycoside-modifying enzymes add bulky chemical groups to the exposed hydroxyl (–OH) and amino (–NH2) groups of the antibiotic, thus preventing it from binding to its target.
2.1.1 Modifying the antibiotic target