1.2.1 Epidemiological cut-off (ECOFF)
The ECOFF is defined as the highest MIC value of isolates that are not known to have resistance and are therefore considered representative of wild type bacterial isolates. Bacteria are defined as ‘wild type’ in the absence of a detectable acquired or mutational resistance mechanism to the antibiotic being considered. ECOFFs reflect the ability to detect resistance mechanisms that may, or may not, be clinically significant. As noted above, ECOFFs are distinct from clinical breakpoints. Clinical breakpoints distinguish between isolates where antibiotic therapy is likely to succeed or fail and take into account the antibiotic dose which a patient can safely receive. You will look at these in the next section.
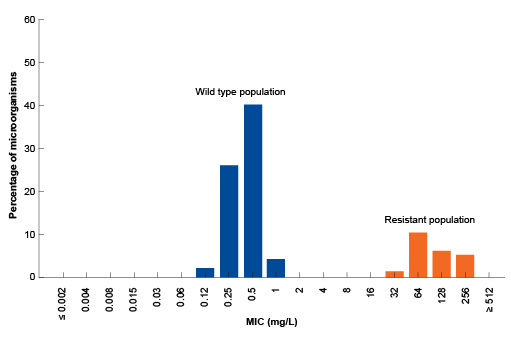
The starting point for defining the ECOFF for a given pathogen-antibiotic combination is an MIC distribution. This gives the MIC values for a number of bacterial isolates that have been extensively characterised and investigated for possible resistance mechanisms. Typically, the wild type MIC distribution spans between three and five two-fold dilution steps, with the ECOFF marking the upper end of the distribution plot (Figures 2 and 3).
What is the ECOFF for the pathogen-antibiotic combination shown in Figure 2?
All the wild type isolates (blue) have an MIC of 1 or less. Therefore, the ECOFF for this pathogen antibiotic combination is 1 mg/L.
In Figure 2, bacteria with detectable resistance mechanisms are clearly distinguishable from the wild type populations. This is because most resistance in this instance is due to a beta-lactamase enzyme made by bacteria. The gene for the enzyme can be acquired by S. aureus, so some isolates may have the gene while some do not. The beta-lactamase enzyme breaks down the amoxicillin, so the antibiotic does not work against the resistant strains. This results in the susceptible bacteria (those that do not produce the enzyme) and resistant bacteria (those that do produce the enzyme) having MIC distributions that are clearly separate.
However, for other pathogen-antibiotic combinations the MIC distribution of bacteria with acquired resistance mechanisms may be closer to, or in some cases overlap with, the wild type MIC distribution (Figure 3).
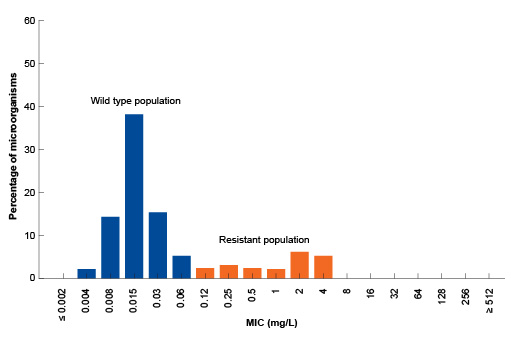
What is the ECOFF for the pathogen-antibiotic combination shown in Figure 3?
The ECOFF for this pathogen antibiotic combination is 0.064 mg/L. The difference between this and the previous example (S. aureus) is because there is a range of variants (mutations) of the penicillin target in clinical isolates of Streptococcus pneumoniae, some of which cause high-level resistance and some of which just raise the MIC a little.
Would an isolate of S. pneumoniae with an MIC for benzylpenicillin of 0.5 mg/L be considered wild type or resistant, according to Figure 3?
It would be considered to be resistant, since the ECOFF for this pathogen is 0.064 mg/L. This means it must be carrying a resistance gene or mutation.
In human and animal health, ECOFF values are mainly used in surveillance rather than to guide therapy. If a clinical breakpoint is available, it should always be used for decisions on treatment rather than an ECOFF value.
Clinical breakpoints are available for most domesticated animal species such as dogs, cats, horses, cattle and pigs. Human clinical breakpoints are often used when veterinary-specific breakpoints are not available. ECOFF values are occasionally used in animal health for some host species where there are few or no clinical breakpoints, for example in aquaculture.
1.2 Interpretive criteria