4.3.1 Enterobacterales species
E. coli and K. pneumoniae are two amongst a variety of coliforms (Enterobacterales).
Your laboratory is likely to be using a differential, indicator medium based on lactose fermentation – usually either
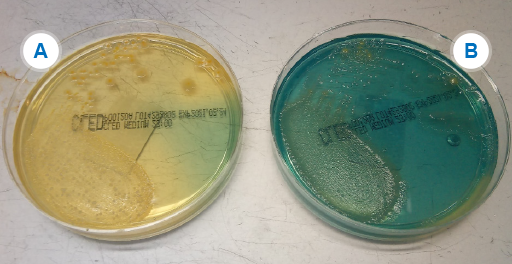
Activity 13: Identifying Enterobacterales in your laboratory
How do you identify the different Enterobacterales species (also known as Enterobacteriaceae or coliforms) in your laboratory? Make some notes and then compare with the sample answer.
Discussion
There are a number of ways of identifying Enterobacterales species depending on budget, number of specimens handled, and relative cost of staffing time versus reagents, available equipment etc.
Methods you may have listed include:
- Indicator media.
- Motility test, biochemical tests (for example, indole test, Voges–Proskauer test etc.)
- Commercial options such as disk forms of the biochemical tests, for example Rosco DiatabsTM or biochemical strip tests, for example
API or Microbact™ - Automated systems or
MALDI-TOF .
The UK Standards for Microbiology Investigations (
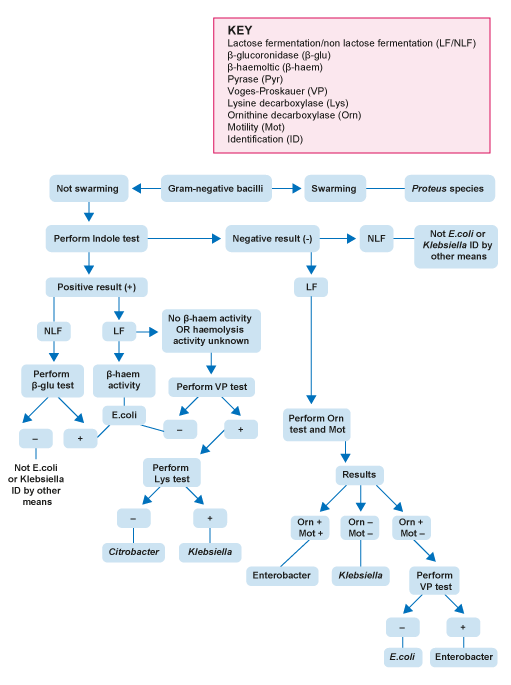
-
Why is it useful to identify Enterobacterales to species level for GLASS?
-
If the species is known, it can make it easier to spot when an organism has deviated from expected patterns of behaviour or developed mechanisms of resistance. For example, a plasmid may have jumped species or the organism acquired new resistance genes. It then becomes possible to focus on tracking and controlling the organism. On a local scale you can identify and control outbreaks quicker.
Resistant organisms of some species are more important pathogens from the Public Health point of view than others. For example, K. pneumoniae can cause big
K. pneumoniae is the only Klebsiella species which is a current focus for
As discussed in Section 3.2 though, only sending the resistant ones to the reference laboratory for species identification could lead to bias in the surveillance figures. This would make it hard to tell the prevalence of the resistant strains in the area accurately. However, it has the major benefit of allowing you to be aware of which resistant Klebsiella species and strains are present in your hospital.
4.3 Identifying Gram-negative organisms



