2.2 Influenza-A and SARS-CoV2
You will now look in a bit more detail at influenza-A and SARS-CoV2, both of which cause acute respiratory infections.
Activity 2 Comparing influenza-A and SARS-CoV2
Take a moment to consider any other similarities. Try to identify three similarities. Write your answer in the box provided below.
Feedback
They are both enveloped viruses, with a ssRNA genome. They are spread by aerosol droplets and you might also have noted that they both cause pandemics, with waves of infection as new strains arise.
Now try to identify three differences between influenza-A and SARS-CoV2. Write your answer in the box provided below.
Feedback
The genome of influenza-A is segmented (eight segments) and negative-sense, whereas that of SARS-CoV2 is a single, positive-sense segment of ssRNA. You might also have spotted that the shape of the viruses is different, as shown in Figure 4.
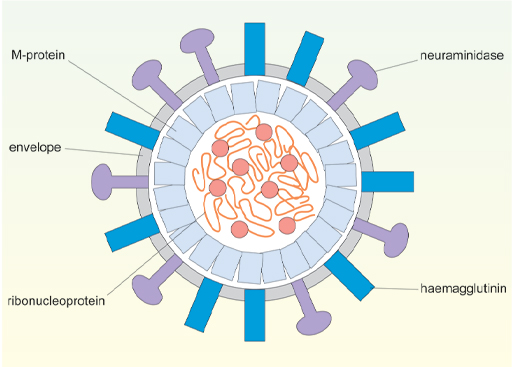
Another difference between these viruses is in how they infect cells and exactly which cells can become infected. The first stage of infection is the binding of a viral surface protein to a protein receptor on the host cell. Since this interaction is highly specific it determines which cell types can be infected by each type of virus. As shown in Figure 5, the nucleocapsid contains viral protein and RNA (ribonucleoprotein) and the envelope of influenza-A has two external viral proteins – the haemagglutinin and neuraminidase.
The haemagglutinin (H) is responsible for attachment of the virus to the target cell. It binds to carbohydrate units (sialic acid) which are attached to a number of different proteins on the host cell surface. (Proteins with bound carbohydrate units are called glycoproteins). Because the target glycoproteins are quite widely distributed on different cell types, influenza-A can infect several different types of cell. The neuraminidase (N) is involved in virus release and spread. Strains of influenza-A are classified according to which haemagglutinin and neuraminidase they have, eg H3N2.
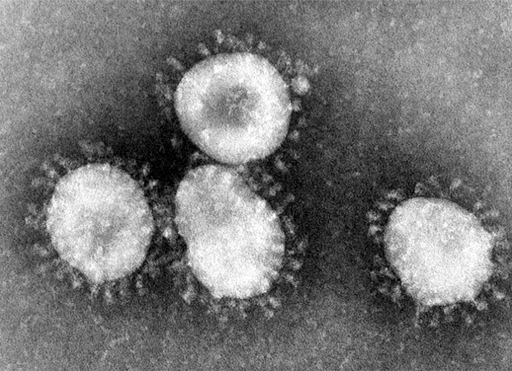
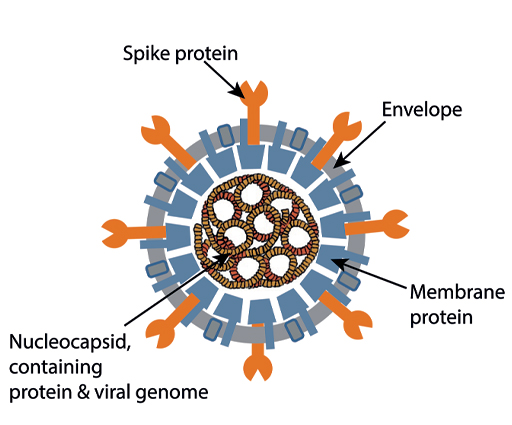
An electron-micrograph of a corona virus is shown in Figure 6. It has prominent spikes on the outside, which give it a crown-like appearance, and this was the origin of the name for this group of viruses. You can see the structure of the virus shown diagrammatically in Figure 7 below.
The spike protein allows the virus to attach to target cells by binding to a protein called the ACE2 receptor (ACE2 = Angiotensin converting enzyme-2). The part of the spike protein which directly contacts the ACE2 receptor is the ‘receptor-binding domain (RBD)’. This detail is very relevant when we consider what antibodies are most effective in protecting against COVID-19 – antibodies which recognise the RBD are particularly important.
-
Would you expect SARS-Cov2, to infect the same cells as influenza-A? If so, why?
-
SARS-Cov2 binds to cells that have the ACE2 receptor, whereas influenza-A binds to a number of surface glycoproteins that are recognised by the haemagglutinin. So we might expect SARS-CoV2 and influenza-A to infect different sets of cells.
Different strains of a virus can also selectively target distinct sets of cells and this is referred to as ‘viral tropism’.
In humans the ACE2 receptor is found in many tissues, but at particularly high levels on the upper surface of epithelial cells facing into the alveoli of the lung and the lumen of the small intestine. It is also found on the endothelial cells which line the inside of blood vessels and in the smooth muscle of arteries. This observation partly explains why the lung is particularly targeted by COVID-19. But other tissues may be affected in more severe cases, when the virus spreads through the body.