1.3 Mutation in the spike protein
As noted previously, mutations tend to cluster in the gene encoding the spike protein, but even there they are not evenly distributed. For example compared with the original Wuhan strain, Omicron BA.1 has a lot of mutations (15) in the receptor-binding domain (RBD), of which five have been shown to enhance binding to the ACE2 receptor. This strain also has a lot of mutation in the N-terminal domain (NTD) , 4 mutations, 3 deletions and one insertion (Figure 2).
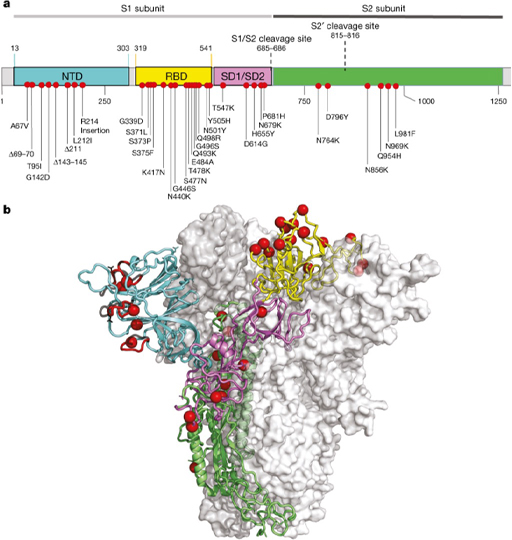
Many of the neutralising antibodies bind to the RBD and NTD regions of the spike protein, so these mutations have allowed the omicron BA.1 variant to substantially evade the antibody response produced by previous variants. Since different people produce antibodies to different regions of the spike protein, the extent to which new variants evade antibodies produced by earlier strains varies between individuals.
