3.1 Evasion of therapeutic antibodies
Recall that monoclonal antibodies recognise just one position (epitope) on an antigen. If a mutation occurs in the epitope recognised by a monoclonal antibody then there is a high likelihood that the antibody will no longer recognise the epitope or bind to it much less well. Also recall that the therapeutic monoclonal antibodies were mostly selected to bind to the RBD of the spike protein, and this is the region that mutates most often.
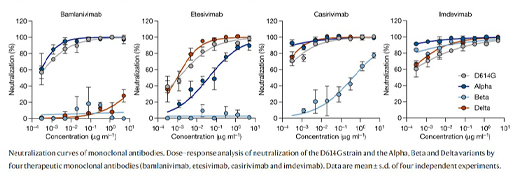
Figure 6 shows neutralisation curves for 4 therapeutic monoclonal antibodies for different SARS-CoV2 variants. The antibodies were raised against the original strain (D614G). In these graphs the concentration of serum is plotted against its ability to neutralise the virus in vitro. If a low concentration of serum causes good neutralisation (curve to the left of the x-axis) then it demonstrates that the antibody is effective against that variant. Notice how the original wild-type strain (D614G) and alpha strains are neutralised by Bamlanivimab, but the beta and delta strains are not. Etesivimab is effective against the D614G and delta strains, partly effective against alpha and ineffective against beta. Casirivimab is effective against D614G, alpha and delta, but has weak activity against beta. Imdevimab is effective against all four strains.
As some of the therapeutic monoclonal antibodies were seen to be ineffective against later virus variants, they were either withdrawn from use, or combined with other monoclonal antibodies, so that the new variant was less likely to be able to evade both of the therapeutic antibodies.
