Unit 3 Final Exam Answer Key
Please only open this once you've completed the final exam.
All correct answers are in red text.
Genetics Unit Exam
Question 1
You are working with an ornamental fish that shows two color phenotypes, red or white. The color is controlled by a single gene. These fish are hermaphrodites – meaning they can either (1) self-fertilize or (2) mate with another fish. You have three fish: fish 1, fish 2, and fish 3.
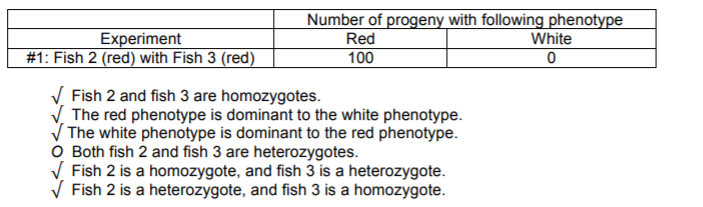
a) You set up the mating experiment #1 below using these fish. From the following statements check all that might be true (that is, are consistent with the data, allowing for reasonable statistical fluctuations), given only the results of experiment #1.
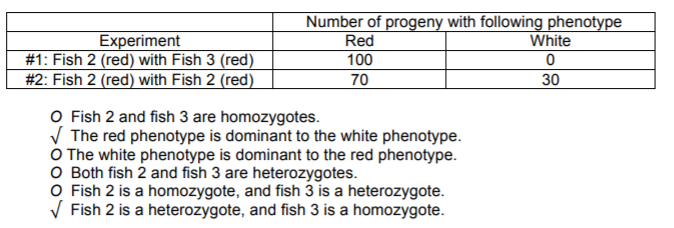
b) You set up mating experiments #1 (same as above) and #2 below using these fish. From the following statements check all that might be true (that is, are consistent with the data, allowing for reasonable statistical fluctuations), given both experiment #1 and experiment 2.
c) You perform experiment #3 and obtain 400 progeny. Note that fish 2 is the same fish in all three experiments. In the table below, list the phenotype or phenotypes you would see in the progeny. Then give the number of offspring expected with each phenotype.

| Possible Phenotypes | Total Number Showing This Phenotype |
|---|---|
| red | 200 |
| white | 200 |
While working further with this fish, you discover one other mutation (in a different gene) that leads to the recessive phenotype of long dorsal fins. We now have two phenotypes:
Color: red or white (described in part a). Alleles denoted by R and r.
Dorsal Fins: long (recessive) or short (dominant). Alleles denoted by D and d
*each trait is controlled by a single gene. For the questions below, use the upper case letter for the allele associated with the dominant phenotype. Use the lower case letter for the allele associated with the recessive phenotype.
In experiment A, you cross true-breeding white fish with short fins to true-breeding red fish with long fins and get all red fish with short fins. You then cross the F1 fish to true-breeding white fish with long fins.
Experiment A: white fish with short fins X red fish with long fins
↓
(F1) red fish with short fins X white fish with long fins
↓
F2: ?
d) You obtain 1000 F2 offspring. Assume that the color and fin loci are 5 cM apart. In the table below, list all the possible phenotypes seen in the F2 offspring. For each phenotype given, list all the possible genotypes seen in the F2 offspring. Finally, give the number of offspring expected with each phenotype.
| Possible Phenotypes | Possible Genotypes | Total F2 Showing this Phenotype |
|---|---|---|
| red fish with short fins | RrDd | 25 |
| red fish with long fins | Rrdd | 475 |
| white fish with long fins | rrdd | 25 |
| white fish with short fins | rrDd | 475 |
Question 2
You are studying a genetically inherited trait. The pedigree for a family with this trait is shown below. This trait shows complete penetrance. Assume no new mutations.
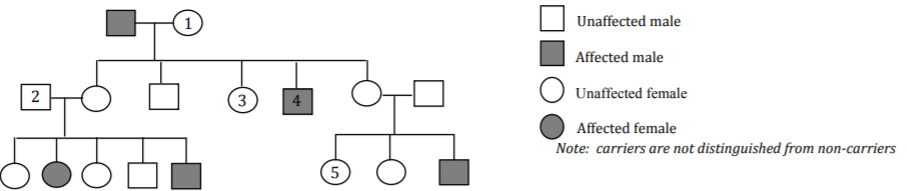
Complete each table below ONLY if that mode of inheritance is possible.
a) Could this trait be inherited in an Autosomal Dominant manner? NO
If Yes, give all the possible genotype or genotypes for the individuals listed in the table below. Use A for the allele associated with the dominant phenotype and a for the allele associated with the recessive phenotype.
| Individual | Possible Genotype(s) |
|---|---|
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 |
b) Could this trait be inherited in an Autosomal Recessive manner? YES
If Yes, give all the possible genotype or genotypes for the individuals listed in the table below. Use A for the allele associated with the dominant phenotype and a for the allele associated with the recessive phenotype.
| Individual | Possible Genotype(s) |
|---|---|
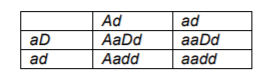
| 1 | Aa |
| 2 | Aa |
| 3 | Aa |
| 4 | aa |
| 5 | AA or Aa |
c) Could this trait be inherited in an X-linked Recessive manner? NO
If Yes, give all the possible genotype or genotypes for the individuals listed in the table below. Use XA for the allele associated with the dominant phenotype and Xa for the allele associated with the recessive phenotype.
| Individual | Possible Genotype(s) |
|---|---|
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 |
Question 3
The pathway for the breakdown of phenylalanine is as follows. There are six enzymes involved in this pathway. Enzyme A is encoded by gene A, enzyme B is encoded by gene B, etc. The genes involved in this pathway all lie on different chromosomes and are thus unlinked to each other.
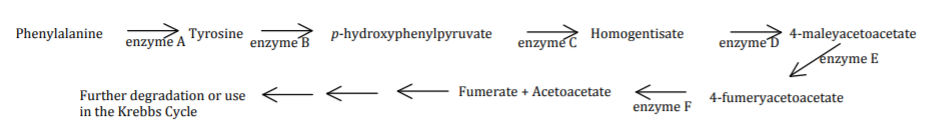
a) PKU is a human autosomal recessive genetic disorder where the inability to breakdown the amino acid phenylalanine and the resulting accumulation can cause neurological problems. What would be the genotype of an individual with PKU? Consider only the relevant gene and use the uppercase letter for the allele associated with the dominant phenotype, the lower case letter for the allele associated with the recessive phenotype.
Genotype of individual with PKU = aa
b) Alkaptonuria is a human autosomal recessive genetic disorder where the individual is missing the function of enzyme D. This results in the accumulation of a byproduct that is excreted in the urine. What byproduct accumulates in these individuals?
Homogentisate accumulates.
c) A woman with PKU who is also a carrier for Alkaptonuria marries a man with Alkaptonuria who is also a carrier for PKU.
i) What would be the genotype of the woman? aaDd
What would be the genotype of the man? Aadd
ii) What are the chances that their child will appear phenotypically normal? Explain your answer or show your work.
25% will be AaDd. (see below) because both traits are recessive, these children will appear normal.
iii) What are the chances that their child will show a both the PKU and the Alkaptonuria phenotype? Explain your answer or show your work.
0%. Although 25% will have the genotype aadd. (see below) these children will show only the PKU
phenotype because the PKU phenotype is epistatic to the alkaptonuria phenotype.
Question 4
You are studying development in a new species of fruit fly, Drosophila mit-ensis, and you find a fly that has curly wings as seen here:

a) The Curly wings phenotype is due to a mutation at a single locus. Assume that you have flies that are true breeding for the curly wing phenotype (cur/cur) and flies that are true-breeding for normal wings (+/+).
P0: cur/cur X +/+
F1: cur/+
• Propose a genetic cross to determine whether the mutant phenotype is dominant or recessive, and list the genotype(s) of the F1 offspring you would obtain from this cross. Use cur for the allele associated with curly wings and + for the allele associated with normal wings.
• What phenotype(s) would you expect to see in the F1 generation if the mutant phenotype is dominant? In what proportion would you see these phenotypes? 100% curly wings
• What phenotype(s) would you expect to see in the F1 generation if the mutant phenotype is recessive? In what proportion would you see these phenotypes? 100% normal wings
You perform the experiment and find that the curly wing phenotype is recessive. A friend of yours has been studying a mutation that causes short antenna. The short antenna phenotype is recessive to the wild-type long antenna phenotype. You designate the mutant allele (ant) and the wild-type allele (+).
You set up the following crosses:
Curly wings, long antenna Normal wings, short antenna
cross 1: cur + + ant
cur + X + ant
⇓
F1: Normal wings,
long antenna
cross 2: Normal wings, long antenna X Curly wings, short antenna
(F1 from cross 1)
b) What type of cross is cross 2? A Test cross
c) If the two genes are unlinked…
i) What phenotypic classes do you expect to see from cross 2 and in what ratios?
Normal wings, long antennae: curly wings, long antennae: Normal wings, short antennae: curly wings, short antennae
1 : 1 : 1 : 1
ii) Could these two genes be located on the same chromosome? Explain.
Yes these two genes can be located on the same chromosome, but far enough apart the recombination
frequency is 50%
d) You perform cross 2, examine 1000 progeny and see that the genes are linked. In the table below, give the genotypes and phenotypes for the parental and recombinant progeny.
Please note that you must use the following nomenclature. Use cur and + for the alleles of the wing gene. Use ant and + for the alleles of the antennae gene.
| Number of Progeny | Genotype | Phenotype |
|---|---|---|
| 431 + 429 | cur + cur ant | Curly wings, long antennae |
| + ant cur ant | Normal wings, short antennae |
| Number of Progeny | Genotype | Phenotype |
|---|---|---|
| 69 + 71 | cur ant cur ant | Curly wings, short antennae |
| + + cur ant | Normal wings, long antennae |
e) What is the recombination frequency between the wing and the antennae genes?
69 + 71/1000 = 14%
Question 5
You are studying a genetically inherited disease. The pedigree for a family with this disease is shown below. Assume 1) the disease phenotype shows complete penetrance and 2) the disease allele is present at a high frequency in the population.
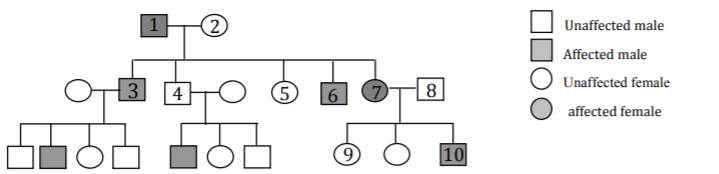
a) Circle all possible modes of inheritance that are consistent with this disease and assumptions 1 and 2.
Autosomal dominant Autosomal recessive X-linked dominant
X-linked recessive Y-linked
*Now assume that individual 8 does not carry any disease alleles. Complete the remainder of this question based on this information.
Use the genotypes +/–, +/+, or –/– for autosomal traits.
Use the genotypes X+ Y, X– Y, X+ X– , X– X– or X+ X+ for X-linked traits.
Use the genotypes XY+ or XY– for Y-linked traits.
*In all cases, + indicates the wild-type allele and – the mutant allele.
b) Give all possible genotypes for the following individuals below.
• individual 2 X+ X-
• individual 4 X- Y