2.4 Infection and replication
Influenza RNA polymerase lacks the ability to recognise and repair any errors that occur during genome duplication, resulting in mistakes in copying its viral RNA about once in every 10 000 nucleotides. Because the influenza genome only contains approximately 14 000 nucleotides, this means that, on average, each new virus produced differs by 1 or 2 nucleotides from its ‘parent’.
The slow accumulation of random genetic changes, especially in the antigenic surface proteins, explains why antibodies that were effective against the virus one year may be less effective against it in subsequent years. This gradual change in the nature of viral antigens is known as antigenic drift.
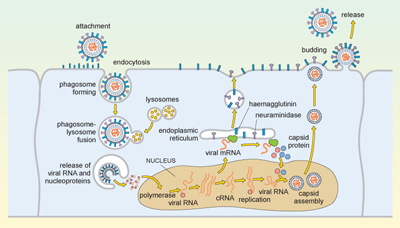
The replication cycle of influenza is illustrated in Figure 5.
Influenza is spread in aerosol droplets that contain virus particles (or by desiccated viral nuclei droplets), and infection may occur if these come into contact with the respiratory tract. Viral neuraminidase cleaves polysaccharides in the protective mucus coating the tract, which allows the virus to reach the surface of the respiratory epithelium.
The haemagglutinin now attaches to glycophorins (sialic-acid-containing glycoproteins) on the surface of the host cell, and the virus is taken up by endocytosis into a phagosome. Acidic lysosomes fuse with the phagosome to form a phagolysosome and the pH inside the phagolysosome falls. This promotes fusion of the viral envelope with the membrane of the phagolysosome, triggering uncoating of the viral capsid and release of viral RNA and nucleoproteins into the cytosol.
The viral genomic RNA then migrates to the nucleus where replication of the viral genome and transcription of viral mRNA occur. These processes require both host and viral enzymes. The viral negative-stranded RNA is replicated by the viral RNA-dependent RNA polymerase, into a positive-sense complementary RNA (cRNA), and these positive and negative RNA strands associate to form double-stranded RNA (dsRNA). The cRNA strand is subsequently replicated again to produce new viral genomic negative-stranded RNA. Some of the cRNA is also processed into mRNA for translation of viral proteins. The infection cycle is rapid and viral molecules can be detected inside the host cell within an hour of the initial infection.
The envelope glycoproteins (haemagglutinin and neuraminidase) are translated in the endoplasmic reticulum, processed and transported to the cell’s plasma membrane. The viral capsid is assembled within the nucleus of the infected cell. The capsid moves to the plasma membrane, where it buds off, taking a segment of membrane containing the haemagglutinin and neuraminidase, and this forms the new viral envelope.
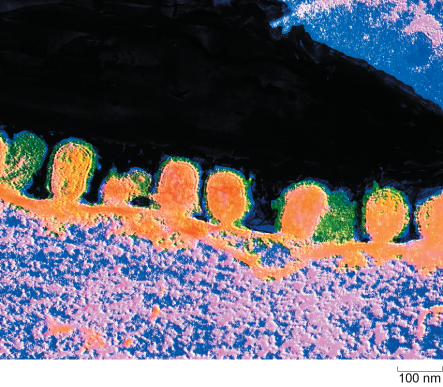
Influenza virus budding from the surface of an infected cell is shown in Figure 6.
From the description above, identify a process or element in the replication cycle which is characteristic of the virus, and which would not normally occur in a mammalian cell.
The replication of RNA on an RNA template with the production of double-stranded RNA would never normally occur in a mammalian cell. Double stranded RNA is therefore a signature of a viral infection. Significantly, cells have a way of detecting the presence of dsRNA, and this activates interferons: molecules involved in limiting viral replication.