3.2.1 Summary of the response
How does the body act quickly to limit viral spread?
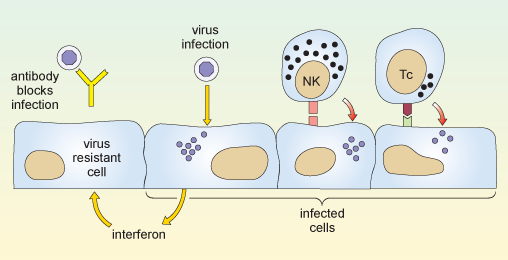
When a virus infects a cell of the body, the molecular machinery for protein synthesis within the cell is usurped as the virus starts to produce its own nucleic acids and proteins. The cell detects the flu dsRNA and other viral molecules and releases interferons, which bind to receptors on neighbouring cells and cause them to synthesise antiviral proteins. If a virus infects such cells they resist viral replication, so fewer viruses are produced and viral spread is delayed.
Also, in the earliest stages of a virus infection the molecules on the cell surface change. Cells lose molecules that identify them as normal ‘self’ cells. At the same time they acquire new molecules encoded by the virus. A group of large, granular lymphocytes recognise these changes and are able to kill the infected cell. This function is called ‘natural killer’ cell action and the lymphocytes that carry it out are termed NK cells.
Non-adaptive and adaptive responses
The actions of both interferons and NK cells in combating infection by influenza occur early in an immune response, and are not specific for the flu virus. These defences occur in response to many different kinds of viral infection, and they are part of our natural, or non-adaptive, immune responses.
Note that immunologists use the term non-adaptive to indicate a type of response that does not improve or adapt with each subsequent infection. This is quite different to its use in evolutionary biology, where it means ‘not advantageous’. Such non-adaptive immune responses slow the spread of an infection so that specific, or adaptive, immune defences can come into play.
The key features of an adaptive immune response are specificity and memory. The immune response is specific to a particular pathogen, and the immune system appears to ‘remember’ the infection, so that if it occurs again the immune response is much more powerful and rapid. Because an immune response is highly specific to a particular pathogen it often means that a response against one strain of virus is ineffective against another – if a virus mutates then the lymphocytes that mediate adaptive immunity are unable to recognise the new strain.
There are two principal arms of the adaptive immune system, mediated by different populations of lymphocytes. One group, called T-lymphocytes, or T cells (which develop in the thymus gland, overlying the heart), recognises antigen fragments associated with cells of the body, including cells which have become infected. A set of cytotoxic T cells (Tc) specifically recognises cells which have become infected and will go on to kill them. In this sense they act in a similar way to NK cells. However they differ from NK cells in that Tc cells are specific for one antigen or infectious agent, whereas NK cells are non-specific.
The second group of lymphocytes are B cells (that differentiate in the bone marrow), which synthesise antibodies that recognise intact antigens, either in body fluids or on the surface of other cells. Activated B cells progress to produce a secreted form of their own surface antibody. Antibodies that recognise the free virus act to target it for uptake and destruction by phagocytic cells.
Therefore the T cells and NK cells deal with the intracellular phase of the viral infection, while the B cells and antibodies recognise and deal with the extracellular virus.
The two reactions described above are illustrated in Figure 8.
You might ask why it takes the adaptive immune response so long to get going. The answer is that the number of T cells and B cells that recognise any specific pathogen is relatively small, so first the lymphocytes which specifically recognise the virus must divide so that there are sufficient to mount an effective immune response. This mechanism is fundamental to all adaptive immune responses.
Activity 2 Influenza mini-lecture
Some of the themes that we have discussed up to now are presented in Video 1: a mini-lecture on influenza by David Male of The Open University. Watch the videos and then attempt the questions below
- Draw a labelled diagram of the structure of influenza A.
- How do pandemic strains of influenza A come about?
Answer
Question 1
Your diagram should look like Figure 4.
Question 2
Pandemic strains of influenza A normally arise by simultaneous infection of a non-human host (typically poultry or pigs) with two or more strains of influenza A. Reassortment of the eight viral segments from each virus allows the generation of a new hybrid virus type, with a completely novel surface structure that has never been seen before by a host immune system (antigenic shift).
