3 Patterns of disease
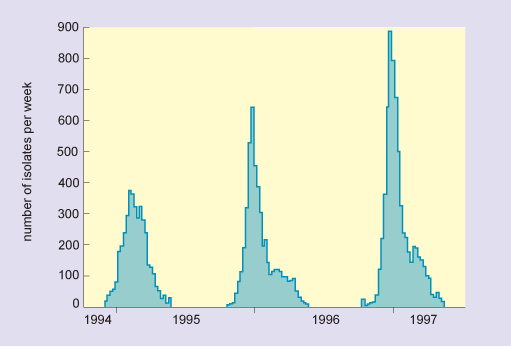
In humans, pigs and horses, flu viruses circulate through populations at regular intervals. The disease is endemic in tropical regions for all of these host groups (i.e. it is continually present in the community). In temperate latitudes, infections are usually seasonal or epidemic, with the greatest numbers occurring in the winter months (Figure 7). Epidemics also occur sporadically in sea mammals and poultry, and in these species high mortality is typical.
In most years, flu in humans affects a minority of the population, the disease course is not very severe and the level of mortality is not great. In such years the influenza virus is slightly different from the previous year due to antigenic drift, which results in the accumulation of genetic mutations that cause the molecules present on the surface of the virus change progressively. In this scenario, the virus is not significantly different from the previous year so that the host’s immune system can more easily mount an effective response than it could to a completely new strain.
However, at irregular intervals the virus undergoes an antigenic shift. This process only occurs in influenza A viruses, typically every 10–30 years, and it is associated with severe pandemics, serious disease and high mortality (see Box 2).
In Section 2.3 you read that strains of influenza are differentiated and designated using a simple system of numbers and letters that depend on their surface antigens. More commonly, however, strains responsible for pandemics are often given a common name according to the area in the world from which they were thought to originate, or the species they mainly affected before becoming transferred to humans (see Table 2). Evidence suggests, however, that in the twentieth century the major flu pandemics all originated in China, with the exception of the 1918 pandemic, which first occurred in the USA.
| Year | Designation | Common name |
|---|---|---|
| 1900 | H3N8 | (none) |
| 1918 | H1N1 | Spanish flu |
| 1957 | H2N2 | Asian flu |
| 1968 | H3N2 | Hong Kong flu |
| 1977 | H1N1 | Russian flu |
| 1997 | H5N1 | Avian flu |
Box 2 The rise of H5N1 – an example of antigenic shift
In 1997, a new strain of influenza A, H5N1, was identified in Hong Kong. The strain was rife in chickens and a few hundred people had become infected. Mortality in these individuals was very high, (6 of 18 died), and so there was serious concern that it marked the beginning of a new pandemic. The authorities in Hong Kong responded by a mass cull of poultry in the region and about 1.5 million chickens were slaughtered. H5N1 did not spread easily from person to person and no further cases were reported in people following the slaughter.
Whether the H5N1 outbreak was an isolated incident of a strain spreading from chicken to humans, or whether it was the start of a major pandemic which was nipped in the bud, cannot be known. Subsequent analysis showed that the high virulence of the new strain could partly be related to the new variant of haemagglutinin (H5), and partly to a more efficient viral polymerase. This outbreak clearly demonstrates the way in which bird influenza can act as a source of new viral strains, and shows that such new strains may be very dangerous to humans.
Since the discovery of the influenza virus in the 1930s it has been possible to isolate and accurately identify each of the epidemic strains, but, as earlier strains of virus have now died out, it has been necessary to infer their identity by examining the antibodies in the serum of affected people.
Antibodies and the ability of the immune system to respond to a strain of flu are much more persistent than the virus itself. It is thus possible to analyse antibodies to determine which types of haemagglutinin and neuraminidase they recognise long after the virus itself has gone. One can then deduce which type of influenza virus that person contracted earlier in their life (as explained in Section 5.2).
