3 Iron uptake by organisms
3.1 How do organisms take up iron?
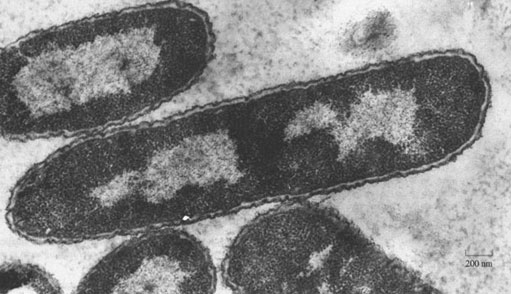
Nearly all organisms are able to take up iron. However, only a handful of organisms have had their iron-uptake chemistry studied. The organism that has received most attention (other than human) is a single-cell, prokaryotic bacterium (found in the human large intestine and elsewhere), called Escherichia coli (abbreviated to E.coli), a high-resolution image of which is shown in Figure 3. The reason that this bacterium has been so thoroughly studied is that it is relatively easy to grow and study colonies of it in the laboratory. The iron-uptake mechanism in E. coli is known in a fair degree of detail.
There are many harmless strains of the E. coli bacterium; the ones found naturally in the human gut are useful because they synthesise several vitamins of the B-complex and vitamin K. However, there are also over 100 pathogenic (i.e. disease-causing) strains of the bacteria. The most infamous is probably E. coli O157:H7, which is very virulent. This strain can find its way into the human food chain (from the intestines of cattle where it is thought to originate), and it causes severe food poisoning due to the toxins excreted by the bacteria. The toxins are absorbed from the gut into the bloodstream; damage to the kidneys occurs, which may eventually result in death, particularly for very old or young persons.
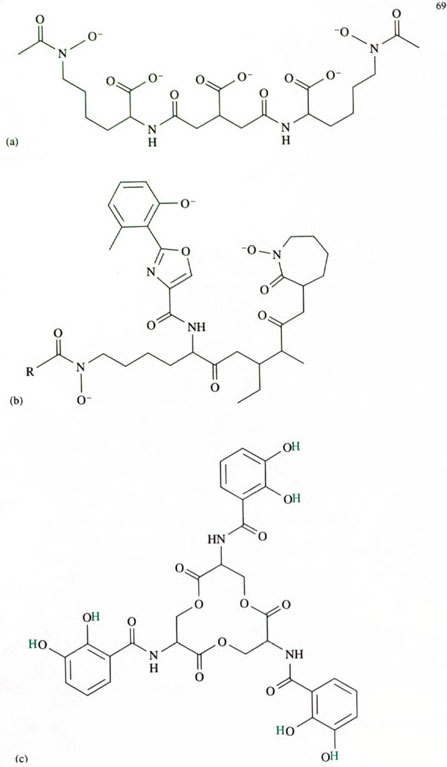
E. coli obtains its iron in a remarkable fashion. Each E. coli bacterium within a colony, secretes small molecules that are capable of specifically chelating iron. These small molecules are known as siderophores (from the Greek for iron carriers; pronounced ‘sid-air-o-fores’). Several types of siderophore are known, each capable of chelating iron in a stable iron–siderophore complex. The structures of some known siderophores are shown in Figure 4.
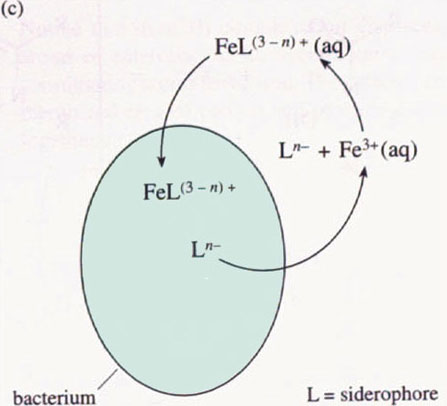
We shall examine the properties of one of the siderophores in more detail below. For all the siderophores, however, their modus operandi is to be secreted from the bacterium, to chelate an iron(III) ion selectively in a stable complex, and then to be re-absorbed by a bacterium (not necessarily the original bacterium) as the iron(III)–siderophore complex (see Figure 5 for a schematic representation).
Activity 11
Derive an expression for the concentration of an iron(III)–siderophore complex in terms of its stability constant. Explain why an iron(III) siderophore complex needs to have a very high stability constant in order to be biochemically useful.
Answer
By writing the equation for the formation of the complex, we can then derive an expression for the concentration of an iron(III)–siderophore complex in terms of its stability constant:
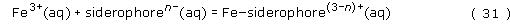
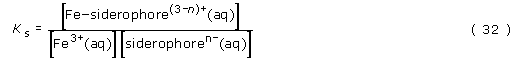
Rearranging gives:
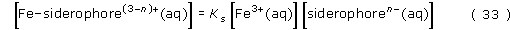
There are two reasons why a high stability constant is required for efficient transport of iron to a bacterium:
Firstly, the value of [iron(III)–siderophore(3−n)+(aq)] must be significant; this is because the bacterium will have a much better statistical chance of absorbing the iron(III)–siderophore complex if it is in relatively high concentration.
Secondly, knowing that iron(III) is in short supply and its concentration in water at pH 7 is very low, any organism that can competitively chelate the available iron will have a better chance of survival.
We have shown that the greater the stability constant, the higher will be the concentration of the iron(III)–siderophore complex, thus providing the optimum conditions for the transport of the iron. Let's take a look at the rough values of [Fe3+(aq)] and [siderophoren−(aq)]. We know that [Fe3+(aq)] cannot be high and may be as low as 10−18 mol l−1, due to the insolubility of many iron(III) compounds. Also, the value of [siderophoren −(aq)] cannot be high (that is, probably much less than 10−12 mol l−1), since the bacterium can only ever synthesise a small amount of the siderophore. What this all means is that for the value of [Fe–siderophore(3 − n)+(aq)] to be significant, the value of the stability constant, Ks must be extremely high. In other words, for this system to be feasible the equilibrium for the complex formation must lie very heavily to the right.
Another requirement of the siderophore ligand is that it must be selective for iron(III), This means that the stability constant for the iron(III)–siderophore complex must be much greater than the stability constants of the siderophore complexes with other metals (including iron(II)). Why is selectivity so important? Firstly, if the siderophore were not selective for iron(III), high concentrations of other metal ions (Mm+) would easily displace the iron(III) from any iron(III)–siderophore complex, according to the equation
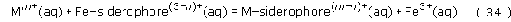
If this equilibrium lay to the right, then iron could not be obtained in any great amounts by the bacterium. Secondly, the bacterium's biochemical systems for absorbing iron should not be a route for the absorption of toxic metal ions, such as mercury and cadmium.
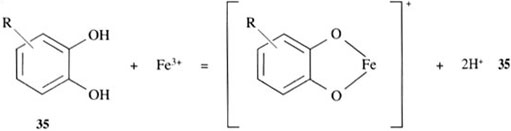
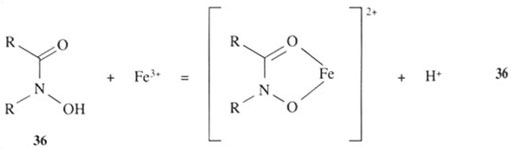
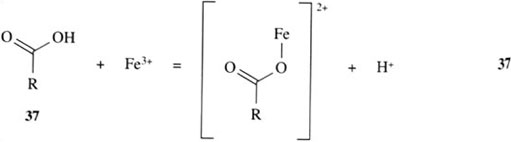
How does a siderophore achieve this high degree of iron(III) selectivity? The question can be partly answered by examining the structures of the siderophores in Figure 4. We can see that the siderophores are analogous to simpler organic molecules which can coordinate directly to iron(III) ion in a similar fashion, as indicated in reactions 35–37:
Activity 12
Are the coordinating groups of the ligands in reactions 35–37 hard or soft?
Answer
All the groups shown are hard ligands and, as such, form stable complexes with hard metals, like iron(III) and aluminium(III).
The groups in structures 35 and 36, 1,2-dihydroxybenzene (trivial name catechol, pronounced ‘kat-a-kol’) and hydroxamic acids, respectively, form particularly stable complexes with iron(III), because they are chelating groups. The chelate ‘bite’ of these two groups is just about right to form a very stable complex with iron(III).
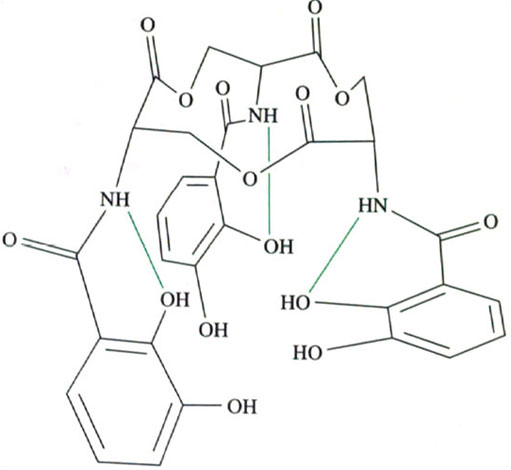
Of all the siderophores, the one that has received most attention is enterobactin, shown in Figure 4c. The reason for this attention is that enterobactin forms an exceedingly stable complex with iron(III); in fact, it is the most stable, soluble iron(III) complex that is known. The stability constant of fully deprotonated enterobactin with iron(III) is extremely high at about 1049 mol−1 l!
Note that the definition of stability constant assumes an equilibrium reaction in aqueous solution between a hydrated metal ion and ligand(s), such that we would write enterobactin in its ionised – that is, fully deprotonated – form.
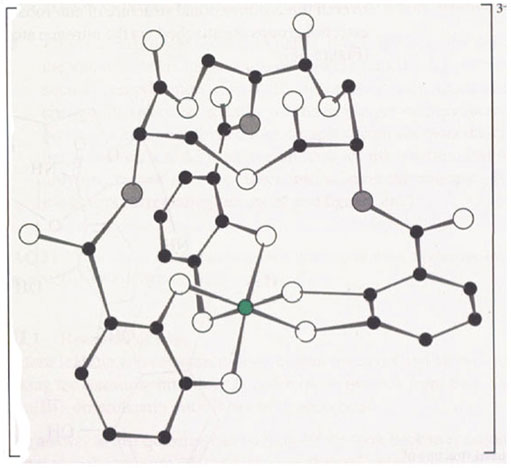
What are the chemical and structural features of enterobactin that give it such a high stability constant with iron(III)? To answer this question we need to examine the structure of enterobactin in more detail. Figure 6 shows the structure of the iron(III)-enterobactin complex.
Activity 13
Which type of group found in siderophore model compounds coordinates to the iron in the complex?
Answer
The iron atom is chelated by three, deprotonated catechol (known as catecholate) groups.
Notice that iron(III) complexation displaces the six protons on the catechol oxygen atoms of enterobactin, so, overall, enterobactin is a hexadentate ligand providing six coordinating atoms to the iron. The catechol rings are attached to each other via a twelve-membered ring of carbon and oxygen atoms. This ring is a serine trimer, condensed together as follows:
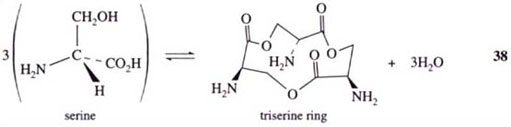
Rather than forming the normal peptide C(O)—NH bond between the individual amino acid molecules, the serines are linked via ester bonds, whereby the ester is formed between the —OH of one serine side-chain and the —CO2H group of another serine:
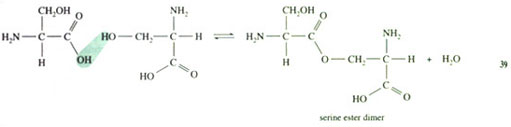
The result is a twelve-membered ring (known as a triserine ring), with three NH2 groups pointing away from the ring. These NH2 groups are all pointing to the same side of the imaginary plane formed by the triserine ring (this is because natural serine exists as a single enantiomer). To complete the enterobactin structure, three catechol groups are attached to the NH2 groups of the triserine ring via amide, C(O)—NH, linkages. The overall three-dimensional structure of enterobactin shows a triserine ring to which three catechol groups are attached via the nitrogen atoms, all linked to the same side of the ring (Figure 7),
Figure 7 also shows that there are other interactions. These interactions are three hydrogen bonds between the NH of a serine group and the oxygen atom of a catechol ring (shown as green lines in Figure 7). This imposes further structural rigidity by preventing each catechol ring from rotating freely, so that not only are the catechol groups all linked to the same side of the triserine ring, but all the calechol oxygen atoms face towards the centre of the ligand.
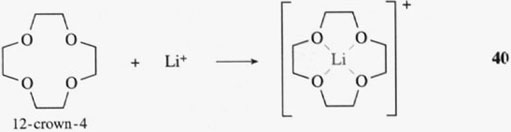
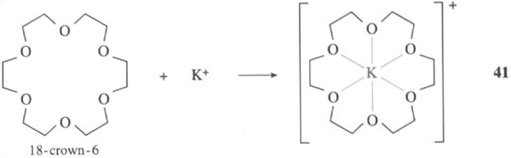
We see that the free enterobactin ligand is actually rather rigid in its structure, with all six coordinating oxygen atoms of the catechol groups held in position to bind an iron(III) ion (that is, the catechol groups have the same relative positions both before and after the binding of iron(III)). This rigid arrangement of functional groups before the metal has been chelated is known as ligand preorganisation; in other words, the three-dimensional structure of the ligand hardly changes on complex formation. What this means in practice is that a metal ion of a particular size (and charge) forms a particularly stable complex with the preorganised ligand. Iron(III) has the correct size and charge to form a very stable complex with the preorganised enterobactin ligand. and, therefore, is chelated selectively by enterobactin. This is exactly the same phenomenon as selective chelation of alkali metal ions by different sizes of crown ethers. A small crown ligand, like 12-crown-4 (reaction 40), will form stable complexes with a small metal ion., Li+, whereas a larger crown, such as 18-crown-6, will form stable complexes with a larger metal ion, K+ (reaction 41):
Activity 14
Why does preorganisation of a ligand lead to a high stability constant?
Answer
This is a manifestation of the chelate effect. If ΔS for the reaction is positive, then the value of ΔG is more negative, and the stability constant is larger. In a reaction such as complexation of iron(III) with enterobactin, the hexadentate ligand hardly changes its structure, and this will have a larger entropy increase than a reaction to produce a hexacoordinated iron complex from six monodentate ligands. (Remember, however, that ΔS is an overall term for the reaction, and the ligand structure is only one part of it; it also takes into account the entropy effects due to the water molecules surrounding the metal and ligand, etc.)
SAQ 1
Summarise the features of enterobactin that make it selective for iron(III) and that make the iron(III)–enterobactin complex highly stable.
Answer
There are three key points. Firstly, enterobactin is a hexadentate ligand and so the stability of the complex is enhanced by the chelate effect. Secondly, the groups that coordinate to iron(III) are hard ligands. Thirdly, the enterobactin ligand is preorganised and must be exactly the correct size, shape and charge for iron(III) binding.