6.4 Evasion of the immune response
All the infectious agents that cause disease in humans have had to evolve ways of evading our immune responses. Even the zoonoses which have other hosts, and diseases such as tetanus and cholera with causative agents that live in the environment in a free-living form, have adapted to survive for at least a period in human hosts.
Some pathogens mutate or change their surface molecules so rapidly that they keep ‘one step ahead’ of the immune response, at least for a time; some disguise themselves from recognition by the immune system; and others have developed ways of limiting the effectiveness of the mechanisms directed against them.
Activity 22
Give examples of pathogens that evade immune responses by: (a) antigenic variation; (b) antigenic disguise; and (c) countermeasures against immune effector mechanisms.
Answer
(a) Influenza virus and HIV, malarial parasites and trypanosomes all undergo rapid antigenic variation. (b) Schistosomes ‘cloak’ themselves with host proteins, disguising their own surface antigens; herpes viruses and pox viruses appear to have incorporated host genes into their own genomes, enabling them to produce proteins that inactivate the complement lytic pathway, (c) Mycobacteria synthesise proteins that inhibit fusion between the lysosomes containing destructive oxygen intermediates and the phagosomes in which they enter the host cell; staphylococci and streptococci have receptors for antibody Fc regions, which compete with Fc receptors on macrophages – the bacteria ‘trap’ the antibodies so they cannot opsonise the bacteria for destruction by the macrophages.
Some additional points about antigenic variation are worth considering in the context of vaccine design. One of the greatest challenges to vaccine development comes from pathogens that mutate their surface antigens very rapidly. In many cases, the areas that mutate are those on exposed loops of external proteins. In HIV, for example, mutation of specific parts of the pathogen's structure can contribute to evasion of the immune response without interfering with the structural integrity of the virus. There are specific regions on gp120, the large surface glycoprotein of HIV, which are particular targets for antibody responses (i.e. they are immunogenic). These regions are particularly susceptible to mutation, so their shapes ‘drift’ as the antibody response builds up, and new clones of B cells have to be activated to cope with the change. However, these immunogenic regions are not vital functional areas of the gp120 molecule. As the gp120 molecule is required by the virus for attachment to CD4 on helper T cells and macrophages, it cannot mutate randomly since it must always retain its ability to bind to CD4. It is notable that the areas that mutate the most are outside the CD4-binding site.
A different kind of problem is seen in trypanosomes, which switch their variant surface glycoproteins. The sole function of these molecules is to protect the outer surface of the parasite and deflect the antibody response. Although immune responses are effective for a time against one VSG, they are ultimately ineffective in controlling the progression of the disease. There are invariant proteins on the trypanosome surface, but these are much less prevalent and less immunogenic than the VSGs, so they are useless as components of a vaccine. One of the main aims of vaccine designers, therefore, is to induce immune responses to those segments of critical antigens that are constrained and cannot mutate without the pathogen losing a key function.
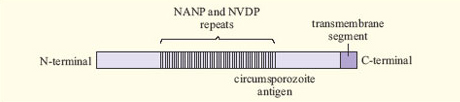
A related problem arises with malaria. The antigens of Plasmodium are extremely complex and vary between different stages of the life cycle of the parasite. Some proteins vary between different Plasmodium strains, while others are relatively constant. An example is the circumsporozoite (CS) antigen, which is involved in attachment of the Plasmodium sporozoite to host liver cells. More than half of the CS protein consists of simple repeats of four amino acids (Figure 10). Such an area may be immunogenic, while being unimportant for protein function.
Activity 23
Explain how other ‘decoy’ proteins of the malaria parasite protect it from host antibody-mediated responses and why they present a challenge for vaccine design.
Answer
Several proteins act as decoys by detaching from the parasite's surface or from the surface of infected red cells. Antibodies against these proteins do not direct an immune response against the parasite itself, and they are ‘mopped up’ by binding to the decoy proteins. Including these proteins in a vaccine would induce the production of antibodies that were similarly ineffective.
The existence of all these escape mechanisms means that it takes a considerable time for even partial immunity to malaria to develop in a naturally-infected population. Consequently, it has been difficult to identify exactly which immune responses would be effective against the parasite, and what to include in a vaccine that could stimulate protective response mechanisms.
