Molecular and physiological biomarkers
The increasing amount of knowledge generated by genome (the catalogue of DNA/genes in a biological tissue), transcriptome (the RNA catalogue) and proteome (the proteins catalogue), sequencing programs for different aquaculture produced fish species, allows us to have an increasingly wider perspective on how nutritional factors influence gene expression and regulation.
The growing number of molecular tools allow the identification of new target genes/proteins involved in nutrient metabolism and a better prediction of a species response to different diets. High-throughput genomic/transcriptomic applications combined with quantification methods are increasingly important for a detailed understanding of the metabolic pathways and the effects of nutritional factors that regulate how fish use their feeds, and also on its consequences on fish health.
Such molecular approaches were also used in the GAIN project to evaluate how aquafeed formulations based on emergent alternative ingredients affected the immune and antioxidant status of Atlantic salmon and gilthead seabream. In a study with gilthead seabream, a transcriptomic analysis with customized PCR-arrays of selected markers showed that 16 out of 44 genes were affected in the liver, 2/29 in the head kidney (HK), and 4/44 in the posterior intestine (PI), when an alternative diet was feed compared to a traditional diet. This allowed us to conclude that the alternative diet leads to positive effects, in the form of a reduced risk of oxidative stress, and a slight anti-inflammatory effect:
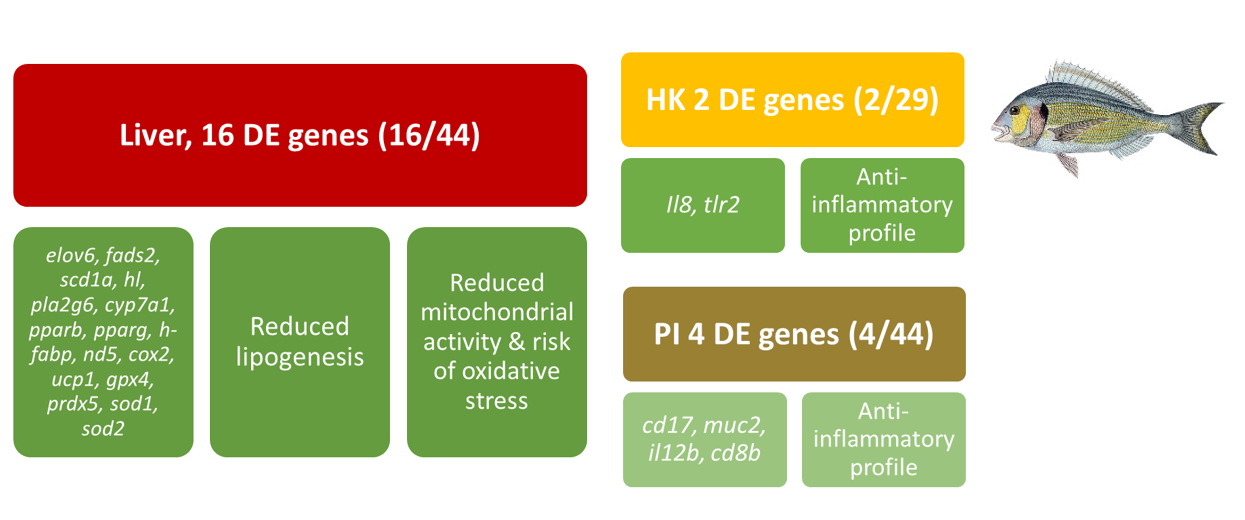
Figure 3. Summary of comparative transcriptomic analysis with customized PCR-arrays of selected genes in the liver, head kidney (HK), and posterior intestine (PI) of gilthead seabream fed with an alternative feed compared to a traditional diet.
Other physiological biomarkers such as enzymes involved in intermediary metabolism (e.g., transaminases) and digestion (e.g., pepsin, trypsin, lipases, and amylase) also provide indications on how fish are using their diets. Alterations in their activities may point to malnutrition or the presence of anti-nutritional factors (ANFs) in the diet.
For a more complete overview of the use of such molecular and physiological biomarkers on aquafeed evaluation please read:
https://www.arraina.eu/images/ARRAINA/Media_Center/Arraina_booklet-V6.pdf