1.8 Protein–protein interactions in signal transduction
Many signalling proteins have both a catalytic domain and sometimes several binding domains.Some only have binding domains, enabling their proteins to act as adaptor, scaffold or anchoring proteins to bring other proteins together. Because of this multiplicity of binding domains, signalling proteins can potentially combine to form complexes with many other proteins; these complexes may be either transient (e.g. in response to stimulation by a growth factor), or stable (to target a protein to an appropriate location). However, protein–protein interactions are not random, as the specific interactions between binding domains and their recognition sites will determine the precise route(s) that a signal transduction pathway will take.
Figure 12 shows a hypothetical signalling cascade, drawn to illustrate how different protein domains have specific functions that result in an ordered network of consecutive protein–protein interactions – in other words, in a signal transduction pathway. Receptor activation by an extracellular signalling molecule leads to the phosphorylation of tyrosine residues on the receptor and of inositol phospholipids on the cytosolic face of the plasma membrane , thereby creating temporary docking sites for an array of SH2- and PH-containing signalling proteins. A cytosolic signalling protein (shown as signalling protein X) contains three different binding domains plus a catalytic kinase domain. On stimulation by an extracellular signalling molecule, signalling protein X translocates to the plasma membrane by virtue of interactions between its SH2 domain and a phosphorylated tyrosine on the receptor protein (sometimes referred as phosphotyrosine or pY), and between its PH domain and phosphorylated inositol phospholipids in the cytosolic leaflet of the lipid bilayer. This translocation results in a change of conformation in protein X, which unfolds a PTB domain, allowing it to bind a phosphorylated tyrosine in protein Y. The kinase domain in signalling protein X then phosphorylates signalling protein Y on another tyrosine, which subsequently binds to the SH2 domain of an adaptor protein. The SH3 domain in the adaptor protein binds to a proline-rich motif on signalling protein Z. This interaction brings protein Z close to protein Y, such that protein Z is phosphorylated at a tyrosine residue. The signal is then relayed downstream by the activated protein Z.
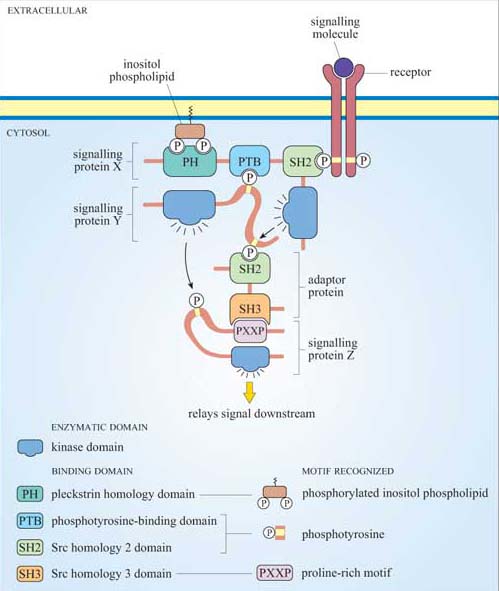
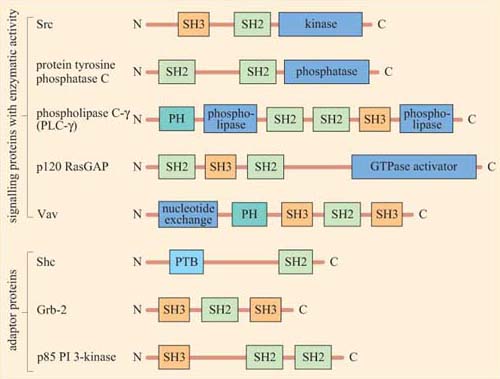
Figure 13 shows the diversity and flexibility of protein-binding domains in some examples of signalling proteins (discussed later in this chapter).
Protein domains can often be identified from their amino acid sequence, and their function deduced from similar, better characterized, proteins. Hence, when a new signalling molecule is identified, it is now often possible to predict, in general terms, from its sequence what it is likely to bind to, and what type of binding domains the signalling molecule contains. It is important to note that whereas the function of a binding domain may sometimes be predicted by the sequence (SH2 domains always bind phosphorylated tyrosines), protein–protein interactions are highly specific — that is, not all phosphorylated tyrosines are recognized by a particular SH2 domain.
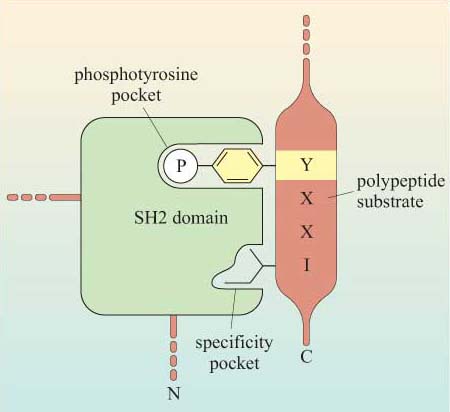
The selectivity of recognition of a motif by a binding domain such as SH2 is conferred by the amino acid sequence adjacent to the phosphorylated residue. We shall illustrate this principle with the SH2 domain of, the tyrosine kinase Src, which has both SH2 and SH3 domains, and a kinase domain (Figure 14a). The core structural elements of its SH2 domain comprise a central hydrophobic antiparallel β sheet, flanked by two short α helices (Figure 14b), which together form a compact flattened hemisphere with two surface pockets. The SH2 domain binds the phosphotyrosine-containing polypeptide substrate via these surface pockets (Figure 15). One pocket (phosphotyrosine pocket) represents the binding site for phosphotyrosine, whereas the specificity pocket allows interaction with residues that are distinct from the phosphotyrosine, in particular the third residue on the C-terminal side of the phosphotyrosine. So, for example, the SH2 domain of Src recognizes the sequence pYXXI, where X is a hydrophilic amino acid, I is isoleucine and pY is phosphorylated tyrosine. Note that all proteins that contain this sequence of amino acids are putative binding partners for the SH2 domain of Src, including the C-terminal phosphotyrosine (pY 527) of Src itself.
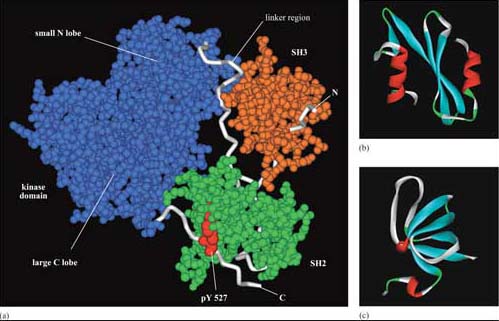
The SH3 domain has a characteristic fold consisting of five β strands, arranged as two tightly packed antiparallel β sheets (Figure 14c). The surface of the SH3 domain bears a flat, hydrophobic ligand-binding pocket, which consists of three shallow grooves defined by aromatic amino acid residues, which determine specificity. In all cases, the region bound by the SH3 domain is proline-rich, and contains the sequence PXXP as a conserved binding motif (where X in this case is any amino acid).
There are various ways of assaying whether signalling proteins interact with each other through their binding domains such as co-immunoprecipitation, yeast two-hybrid screening, proteomics and FRET. Box 2 describes another technique used to analyse protein–protein interactions that you will use in Experimental investigation 3 at the end of this chapter.
Box 2 Use of fusion proteins for pull-down assays in the study of signalling protein domain interactions
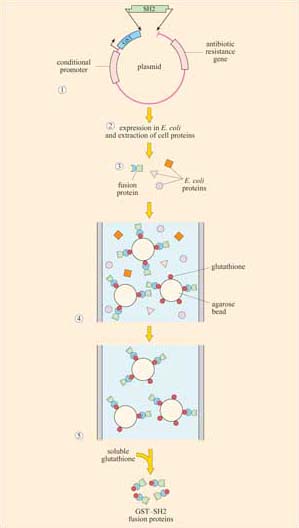
Individual domains often retain their function when isolated from their parent protein, and they can be genetically engineered to be fused with other proteins/peptides. Recombinant fusion proteins consist of two proteins: (a) a protein or peptide sequence used as a tag to facilitate protein isolation; and (b) the ‘bait’ protein, used as a means of indirectly ‘pulling down’ interacting proteins. One very useful example of this technique is the use of glutathione S -transferase (GST) fusion proteins; they consist of GST (tag) fused to a protein or part of a protein of interest (bait).
Using recombinant DNA technology, the DNA encoding the domain of interest is inserted into a plasmid vector just downstream of, and in the same translation reading frame (ORF) as the gene for GST. Under optimized conditions, certain strains of bacteria are induced to take up the plasmid and grown in selection media. Expression of the fusion protein is then chemically induced in transformed bacteria, which are subsequently lysed in a detergent solution. GST is used as the fusion partner because it binds glutathione, a property that can be exploited to purify the fusion protein by affinity chromatography. Free glutathione can be used to elute the fusion protein from the column. The GST can then be cleaved from the protein being investigated, if not further required. This gentle technique produces fusion protein of sufficient quantity and quality for use in, for example, binding assays and enzyme activity assays. Figures 16 and 17 show one example of its use.